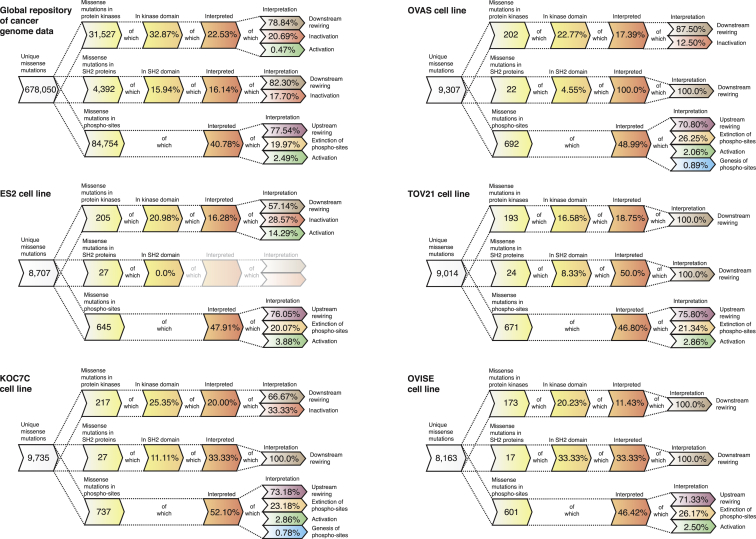
Figure 2.
Overview of NAMs in Cancer Cell Lines and in the Global Repository of Cancer Somatic Mutations as Predicted by ReKINect
For each cell line and for the global repository of cancer somatic mutations we show the number of unique missense variants and how many of these variants fall within kinase proteins, SH2 proteins or phosphorylation sites (using a five-residue flanking region window surrounding the phosphorylation site). From these we then illustrate the fraction of variants falling within the respective domains and the fraction that can be interpreted by ReKINect. In the case of ES2, all of the 27 variants hitting an SH2 protein, hit outside SH2 domains, thus ReKINect could not make any predictions as to their effect (ghosted). It should be noted that the genesis of phosphorylation sites cannot be predicted from in silico analysis alone but require genome-specific-MS experiments. See also Figure S1.