Figure S1.
Interpreting Functional Cancer Somatic Mutations in Repositories of Cancer Genome Data and Cell Lines, Related to Figures 1 and 2
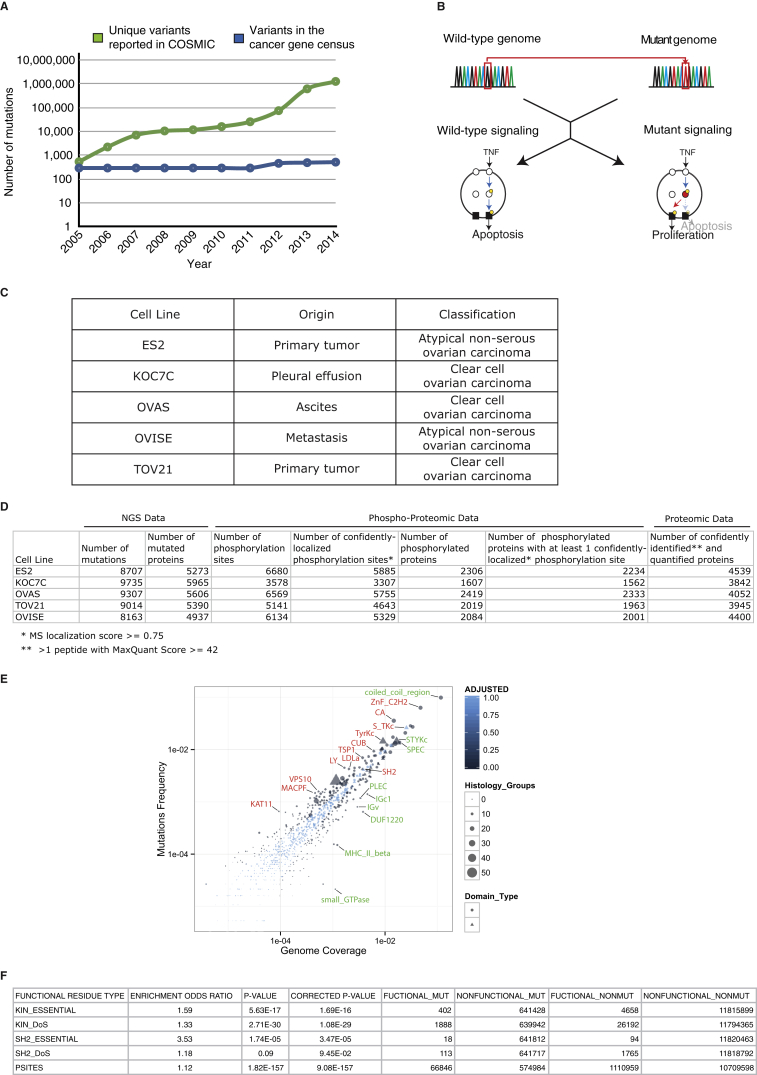
(A) The gap between the number of unique cancer somatic mutations reported by global sequencing efforts and the ones for which the community has been able to attribute a driving role in cancer (list of genes in the cancer gene census [Forbes et al., 2011]) has been growing drastically in the last years.
(B) A more comprehensive understanding of signaling networks and how mutations perturb them would help close the interpretation gap described in (A).
(C and D) Data summary of the provenance (C) and different experimental observations (D) concerning mutations, phosphorylation sites, and proteins found using exome sequencing (NGS) and (phospho)proteomics-based mass spectrometry. Stringent filters were applied to ensure data quality, including 95% of sequence space covered by 10X sequencing reads for the NGS data, standard filters applied in subsequent steps (see Experimental Procedures), and high MaxQuant and localization scores for the MS data (see Experimental Procedures).
(E) Using the global repository of somatic cancer mutations, we quantified the enrichment of mutations in functional residues covered by ReKINect, and to what extent different protein domains are affected by somatic missense mutations. As one would expect, and can be observed in (E), the mutation frequency generally depends on the fraction of the genome that a given domain covers (genome coverage), as shown in the scatter plot. However, several signaling (triangles) and non-signaling (circles) domains harbor many more mutations than it would be expected by random chance or genome coverage alone (darker blue denoting lower P-value) and are mutated in a wider range of cancers (data point size). These include signaling domains like serine-threonine kinase domains, S_TKc, tyrosine kinase domains, TyrKc, and SH2 domains.
(F) The results on (B) show the enrichment in cancer mutations on specific residues, calculated as the fraction of functional residues mutated and not mutated with respect to the fraction of the proteome covered by functional and non-functional residues. Odds-ratios and P-values were computed using a Fisher’s Exact Test with Multiple-Test Correction.