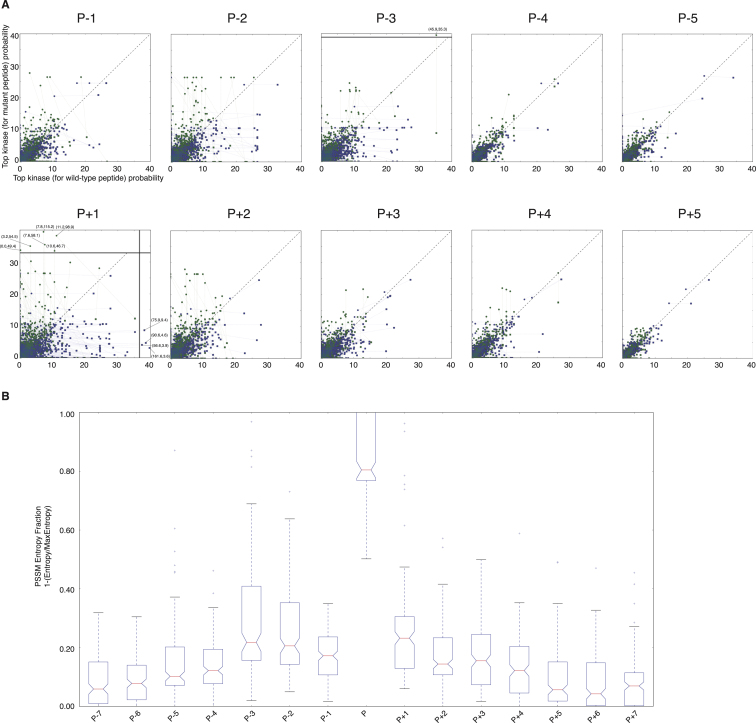
Figure S5.
Upstream Rewiring Graphs Using NetworKIN and Information Content in Various Phosphorylation Substrate Positions, Related to Figure 5
(A) As an extension to the graphs shown in Figure 5, here we show similar rewiring graphs computed using NetworKIN (Linding et al., 2007) instead of NetPhorest (Miller et al., 2008), and therefore including contextual information for improved accuracy. Please note that the extreme values in P-3 and P+1 graphs were added for completeness, but due to their outlier status are out of scale (values added and their numerical values can be seen in the figure); for further information and accurate numerical values please refer to Table S5 online. It is important to note that we only used top-scoring NetworKIN (Linding et al., 2007) and NetPhorest (Miller et al., 2008) predictions filtered to ensure maximum confidence, and that we reached the same conclusions using both algorithms. Because of this and the fact we have based our global observations on thousands of mutations, our conclusions drawn from Figure 5 and (B) are highly unlikely to have been affected by our choice of methods.
(B) By analyzing PSSMs characterizing the peptide specificity of a large number of human protein kinases (from the NetPhorest repository), we could quantify how much each substrate position contributes to the kinase-substrate recognition process (from seven residues before the phosphorylation position, P-7, up to seven residues after, P+7). Similar to what we observed in Figure 5 and (A), in the case of mutations hitting different positions and their likelihood to lead to upstream rewiring, position P-1 contributes relatively little to the kinase-substrate recognition process.