Figure S2.
Related to Figure 2
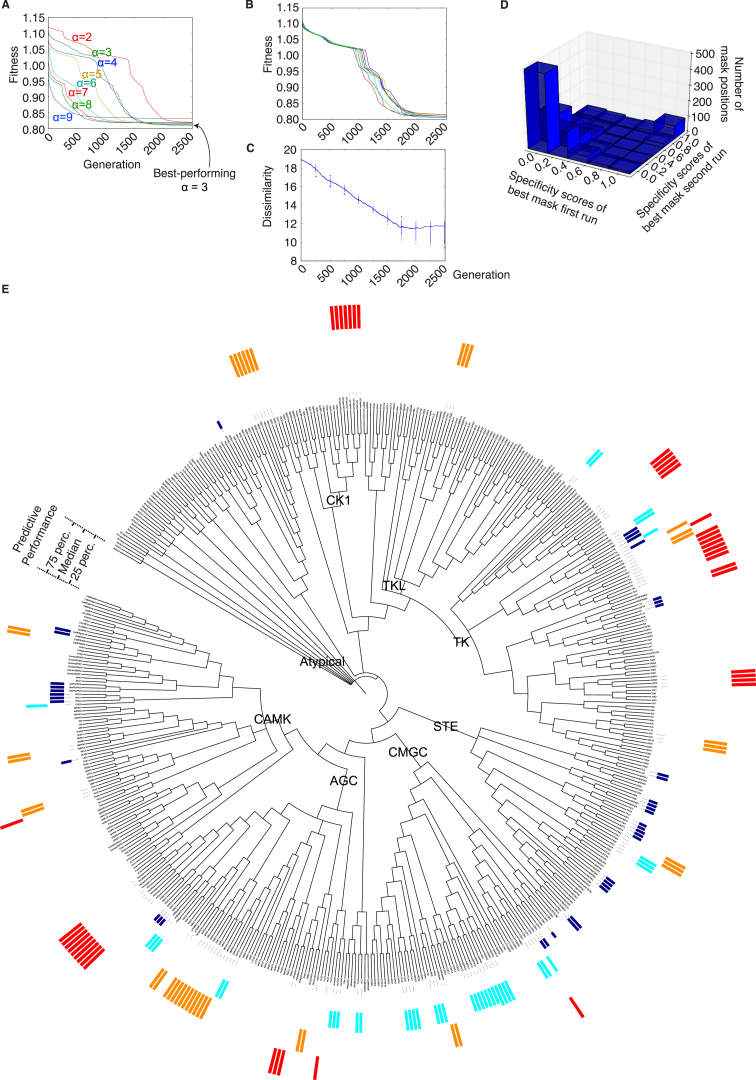
(A) Alpha Determination for Kinase KINspect. As explained in Experimental Procedures, a parameter ‘alpha’ (α) needs to be optimized to determine the best trade-off between using only the most similar domains or include more distant domains when predicting new PSSMs. In essence, the procedure described for KINspect in Figure 2 is performed using different alphas and the alpha leading to the best performance is chosen. As shown here, the best results (lower prediction error) were obtained with α = 3, thus this value was used subsequently. Even though, in line with standard nomenclature for genetic algorithm, we have labeled the y axis as being “Fitness,” it is important to clarify that KINspect evolves by minimization the error in predictions, therefore “minimizing fitness.” This “Fitness” is measured as the median Frobenius distance between predicted and experimentally determined PSSMs.
(B) KINspect fitness trajectories. When trained on the human kinome, KINspect reaches convergence after approximately 2000–2500 generations. Fitness is measured as the median Frobenius distance between predicted and observed PSSMs. Each color in this plot shows the fitness of the best mask at each generation. The similarity between the different trajectories representing the 10 independent KINspect evaluation runs confirms they have followed a similar path to convergence.
(C) KINspect convergence, robustness and performance. In order to evaluate whether similar results are obtained in the 10 independent KINspect evaluations, the best mask for each run is compared to all the others at each generation and their dissimilarity is measured as the Frobenius distance between the vectors. By including box-plots every 500 generations, we could also assess the evolution of the overall distribution. The graph illustrates the increase in similarity (decrease in dissimilarity) of results as one moves closer to the final point of convergence. From this, one can conclude that independent algorithm deployments tend to converge to the same (or at least highly similar) solution. One can further appreciate the similarity corresponding to this Frobenius distance by referring to (C), where the scores of two masks at this distance are represented pair-wise.
(D) By comparing two of the final specificity masks obtained in two independent KINspect evaluations, we could compare the score of the two masks at the same kinase domain positions. This distribution shows a large degree of agreement (e.g., residues scoring 1 in one masks have a high tendency to score 1 in the other one) between the two final masks obtained in two independent KINspect evaluation runs, as well as a strong tendency for most residues to score 0 in both runs.
(E) KINspect coverage. Overview of the predictive performance of KINspect for different human kinase domains. A larger bar indicates higher (better) predictive performance, while a shorter bar indicates lower (worse) predictive performance. For more clarity, bars have been colored in dark, light blue, orange or red (predictive performance below the percentile 25, below the median, above the median or above the percentile 75, respectively).