Figure S4.
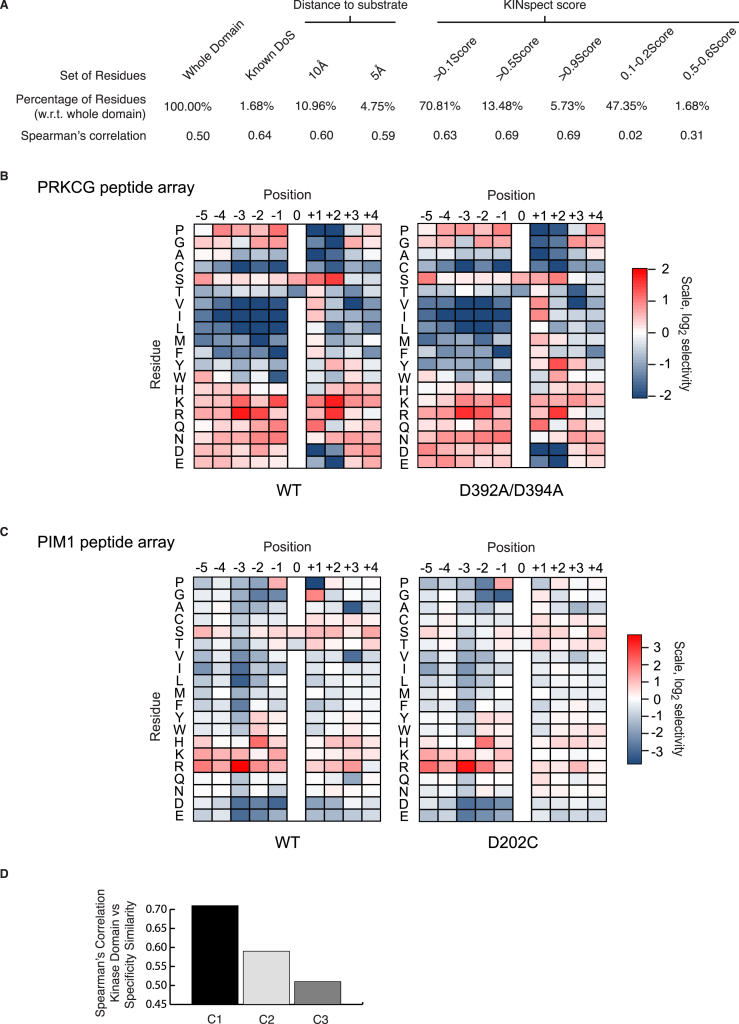
(A) No Other Separation Method Leads to a Higher Sequence-to-Specificity Correlation than KINspect. As shown in the figure, the correlation varies between different subsets of residues, but is highest for residues that obtained high KINspect scores (KINspect Score > 0.9) despite being a relatively smaller fraction of domain alignment positions.
(B and C) Complete PSSMs Obtained by PSPL for Wild-type and Mutant Kinase Experimentally Validating KINspect Predictions. Extended version of the PSSMs represented in Figure 3 showing the changes in peptide preferences when mutating three of the residues with high KINspect score.
(D) Contribution to Specificity From Each Specificity Cluster. In order to quantify whether the different clusters of highly scoring residues contributed equally to specificity we computed the Spearman correlation using only residues belonging to each cluster. From the graph, it can be concluded that Cluster-1 encodes most of the specificity, in line with its closer proximity to and direct contact with the peptide substrate, followed by Cluster-2 and Cluster-3.