Figure S5.
Related to Figure 5
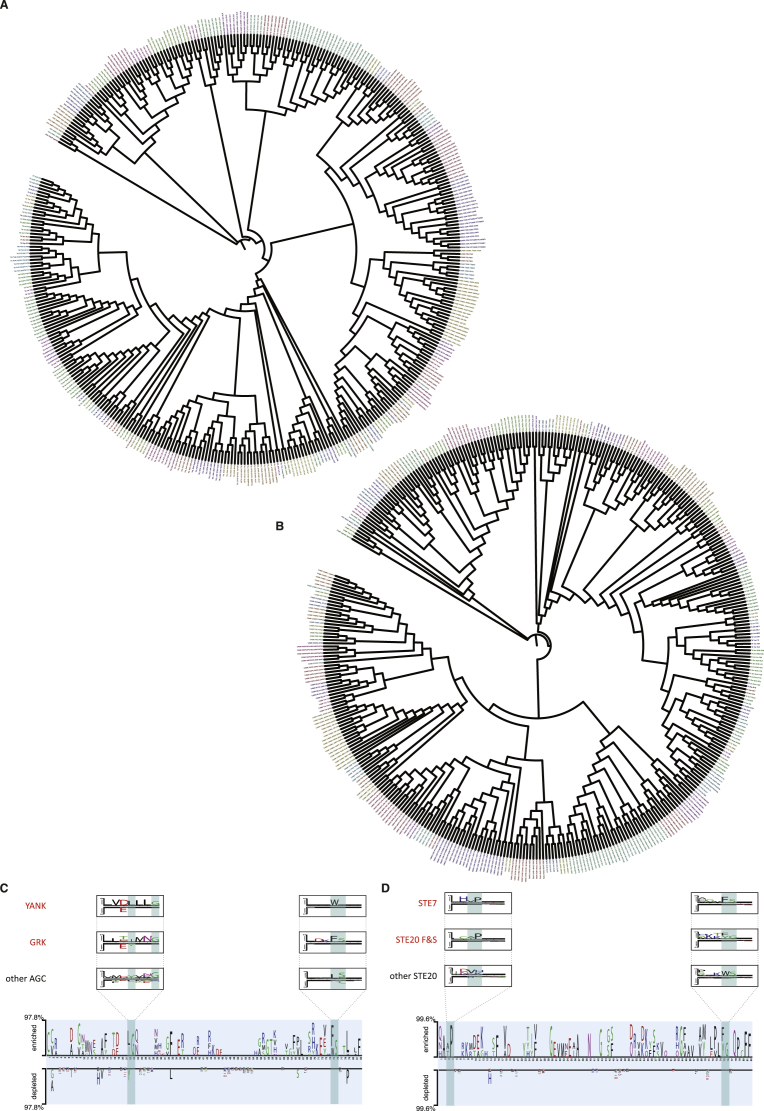
(A) DoS-Based Evolutionary Tree. By producing a sequence alignment including only positions identified as determinants of specificity by KINspect, we could explore how kinase domain specificity evolved (as compared to the evolution of the domain as a whole, B). Several examples of significant differences between the domain-wide and DoS-centric evolutionary trees (as shown in Figure 5) confirm how these two evolutionary paths, of domain overall function and specificity, are not completely coupled. This figure was created with iTOL (http://itol.embl.de; Letunic and Bork, 2007).
(B) Whole-Domain Evolutionary Tree. For comparison purposes, as explained in Figures 5, we built a domain-wide evolutionary tree following the same strategy as in the case of the DoS-based tree, but taking into consideration all residues within the kinase domain.
(C and D) DoS and Motif Logos that Cause Kinase Family Re-arrangements in the DoS-based tree. Further exploration of the DoS and DoS-centered alignment clarifies the reasons why there are marked differences between the DoS-based tree and the one where the whole domain was considered. In the case of Yank and Grk families, for instance, a preference for Leucine (L) over Tyrosine (Y) in the first highlighted DoS and for large hydrophobic amino-acids (as opposed to Leucine) in the other one, exemplify why they cluster separately from other AGC kinases. In the case of the STE7 family embedding within the STE20 family, a couple of strong preferences for Proline (P) and Phenylalanine (F) in the two highlighted DoS provide evidence for the clustering of kinases belonging to two different families (i.e., STE7 and STE20’s STLKs and FRAYs). For further details on how these logos were built please refer to Supplemental Experimental Procedures.