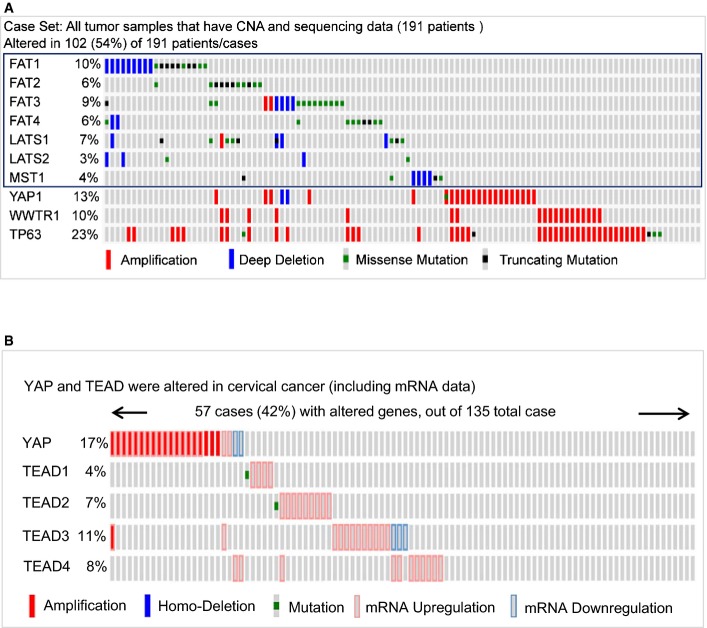
Figure EV1. Multidimensional cancer genomics data analysis showing alteration of the major genes involved in the Hippo/YAP pathway in cervical cancer.
- Alteration frequencies of major genes involved in the Hippo pathway. Genes in the blue box are upstream genes of the Hippo tumor suppressor pathway. Note the frequent deletion and mutation of these genes in cervical cancer. YAP and TAZ (WWTR1) genes are frequently amplified in cervical cancer (n = 191). TP63 are a known cervical cancer biomarker and are frequently amplified in cervical cancer.
- A visual summary of the different mechanisms of YAP1 and TEAD alteration across a set of cervical cancer samples based on a query of the five genes, YAP, TEAD1, TEAD2, TEAD3, and TEAD4. Each row represents a gene, and each column represents a tumor sample (n = 135). The YAP and TEADs gene alteration analyses were performed using online datasets and data mining tools (the cBioPortal for Cancer Genomics and the datasets from the TCGA Research Network).