Figure EV1. Comparison of proteomic workflows for blastocoel analysis and development of targeted proteomic assays for single embryo protein quantification.
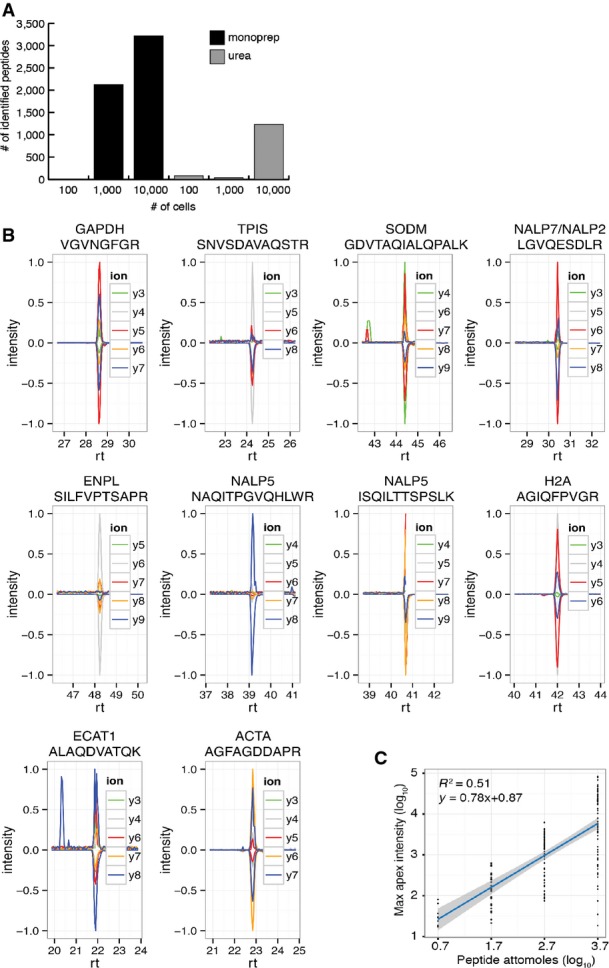
- Number of peptides identified using different sample preparation methods (black: solvent-based, MonoPrep; gray: urea-based, standard) on cytosolic lysates of HeLa cells at different concentrations (100, 1,000 and 10,000 cells corresponding to an estimated protein amount of 8, 80 and 800 ng, respectively).
- Validation of ten SRM assays on samples of five blastocoels. Batched samples obtained from 5 blastocoels were spiked with isotopically labeled versions of the target peptides. SRM assays were manually validated on the basis of the co-elution and relative intensities of at least 4 transitions per assay. Transition traces are displayed using their relative intensities (with the intensity of the highest transition per assay being set to 1). Positive values are used for endogenous peptides, while negative values are used for reference (synthetic) peptides. Horizontal axis displays peptide retention time (rt) in minutes.
- Intensity calibration curve derived from 76 absolutely quantified (AQUA) peptides measured at different concentrations. The linear regression between log10-transformed maximum apex intensity (derived from the maximum of the most intense transition trace) and log10-transformed attomoles of peptide injected was used to transform SRM-derived peptide intensities for blastocoel proteins into absolute abundances.
