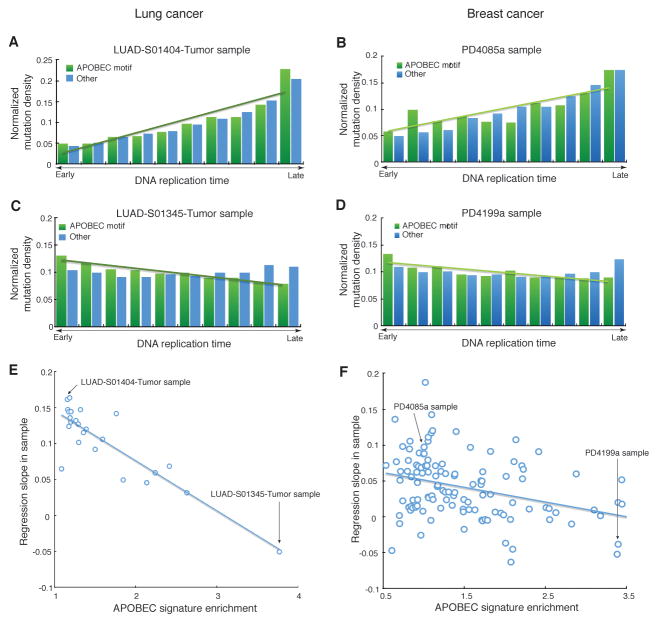
Figure 2. Dependence of the normalized density of APOBEC-signature mutations from replication timing of a cancer genome region.
(A,B) Samples with low or no enrichment with APOBEC mutation signature (fold enrichment <2) display a positive correlation between the normalized density of APOBEC-signature mutations and replication timing (an example for lung cancer is shown in panel A, an example for breast cancer is shown in panel B).
(C,D) Samples with high enrichment with APOBEC mutation (fold enrichment ≥2) display a negative correlation between the normalized density of APOBEC-signature mutations and replication timing (an example for lung cancer is shown in panel C, an example for breast cancer is shown in panel D). Bins on the horizontal axes in A–D panels were obtained by sorting all genome positions by the values of a genomic feature (DNA replication time or chromatin accessibility) and then dividing into ten non-overlapping equal-sized windows.
(E,F) In general, the slopes of this regression are anti-correlated with APOBEC-signature enrichment (lung cancer data is shown in panel E and breast cancer data is shown in panel F). Similar analyses with respect to chromatin accessibility are shown on Figure S2 and analyses performed separately for the two subcategories of APOBEC-signature mutations (TCW→TTW and TCW→TGW) are shown on Figure S3. The sample specific enrichment values used for the analyses in Figure 2 and Figure S3 are shown in Table S1.