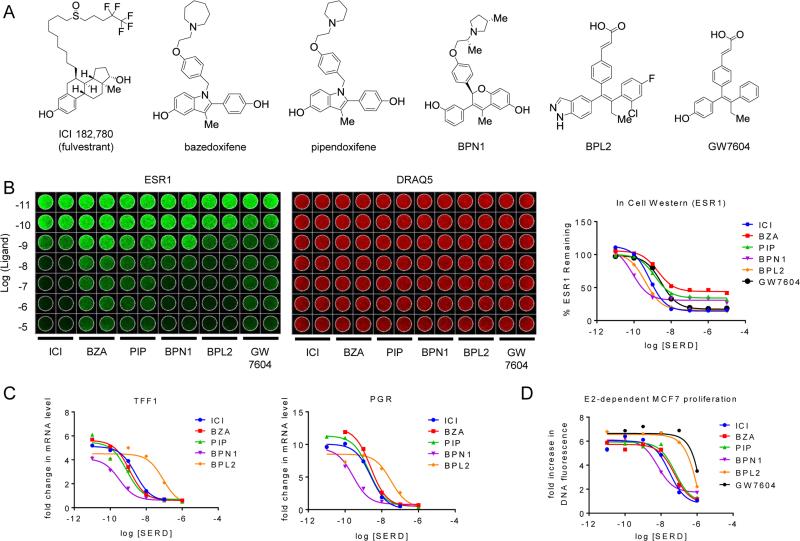
Figure 1. SERDs and SERM/SERD hybrids inhibit ESR1 action with similar efficacy despite differences in potency and efficiency of estrogen receptor turnover.
A) Chemical structures of SERM/SERD hybrids (SSHs) and SERDs evaluated. B) MCF7 cells were incubated with increasing concentrations (10−11–10−5 M) SERD ICI 182,780 (ICI), or SSHs bazedoxifene (BZA), pipendoxifene (PIP), BPN1, or BPL2 for 18 hrs. ESR1 protein levels (left) were assessed using an In Cell Western. Data was normalized to DNA content using DRAQ5 (center) and quantitated (average of duplicate wells) using GraphPad Prism 6 (right). C) MCF7 breast cancer cells were treated for 24 hours with 1 nM E2 in the presence of increasing concentrations (10−11 – 10−6 M) of antagonist. mRNA levels of ESR1 target genes progesterone receptor (PGR) and trefoil factor 1 (TFF1) were assessed using real time quantitative PCR (RT qPCR) following RNA isolation. mRNA expression was normalized to the 36B4 housekeeping gene, and expression levels are presented as fold change as compared to an untreated control. D) MCF7 cells were plated in phenol red free media supplemented with charcoal stripped FBS 24 hours prior to treatment, and were treated with 1nM E2 as well as with the indicated ligands (10−11 – 10−6 M) on days 1, 4, and 6 of an 8 day proliferation assay. DNA content, as assessed by fluorescence (FluoReporter assay), was measured as a surrogate for cell proliferation. The relative increase in DNA fluorescence was calculated by normalizing to baseline values detected in a duplicate plate of cells that was harvested on day 1 prior to the initial treatment. Data are representative of at least 3 independent experiments.