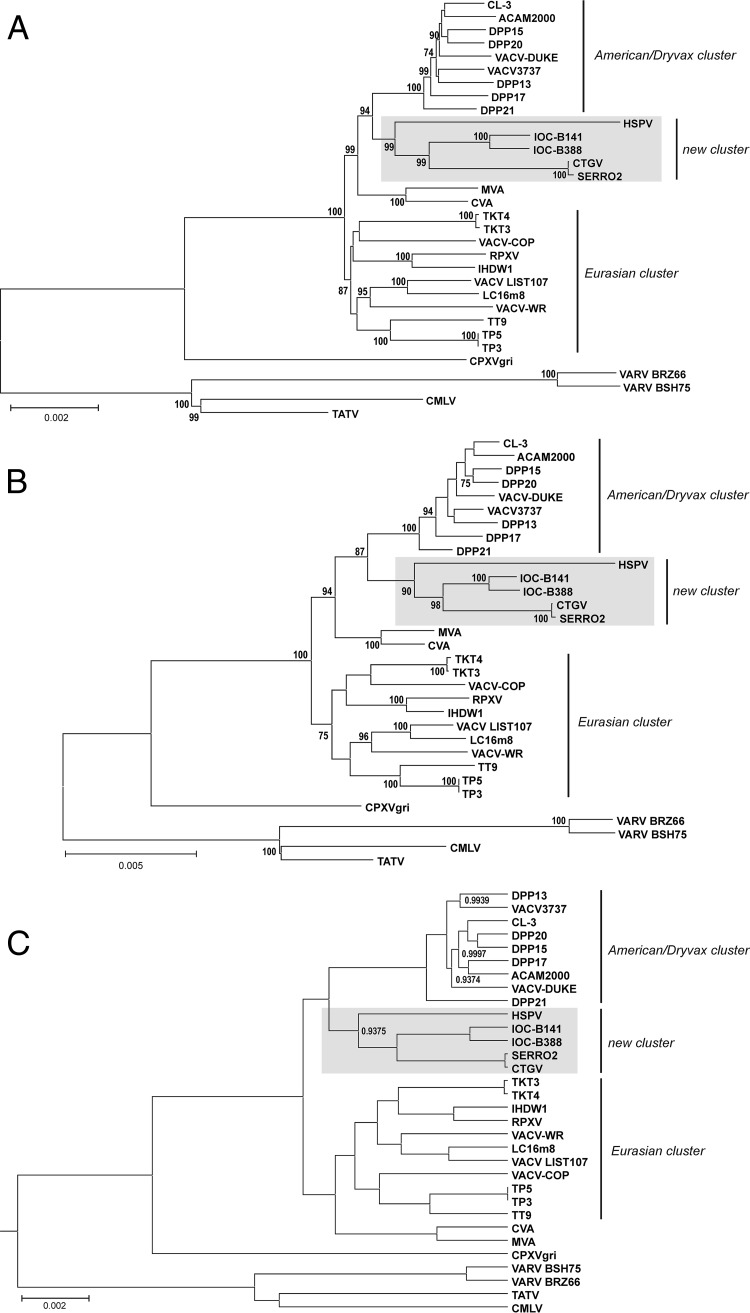
FIG 8.
B141 and B388 branch as a novel cluster in VACV phylogeny. The multialignment of a conserved region from 31 Orthopoxvirus genomes (orthologs of VACV-Cop F9L through A24R) was used for phylogenetic inference. (A) Neighbor-joining (NJ) tree based on Kimura 2-parameter substitution model with 1,000 bootstrap replicates. (B) Maximum likelihood (ML) tree based on the Tamura-Nei substitution model, a five-category discrete gamma model with 1,000 bootstrap replicates. NJ and ML trees were constructed using MEGA 6.06, and bootstrap values of >70% are shown next to branch nodes. (C) Maximum clade credibility tree constructed by Bayesian inference (BEAST v. 1.8.1.) with default prior settings, 4 chains of 10 million generations of Markov chain Monte Carlo (MCMC) with tree sampling every 1,000th generation, and a burn-in of 1,500,000 states. Nodes showing posterior probabilities of <1.00 are labeled. (A, B, and C) The scale bars indicate the number of substitutions per site. The VACV clusters are indicated on the right. Gray boxes highlight the new VACV cluster grouping IOC-B141, IOC-B388, CTGV, Serro 2 virus, and HSPV.