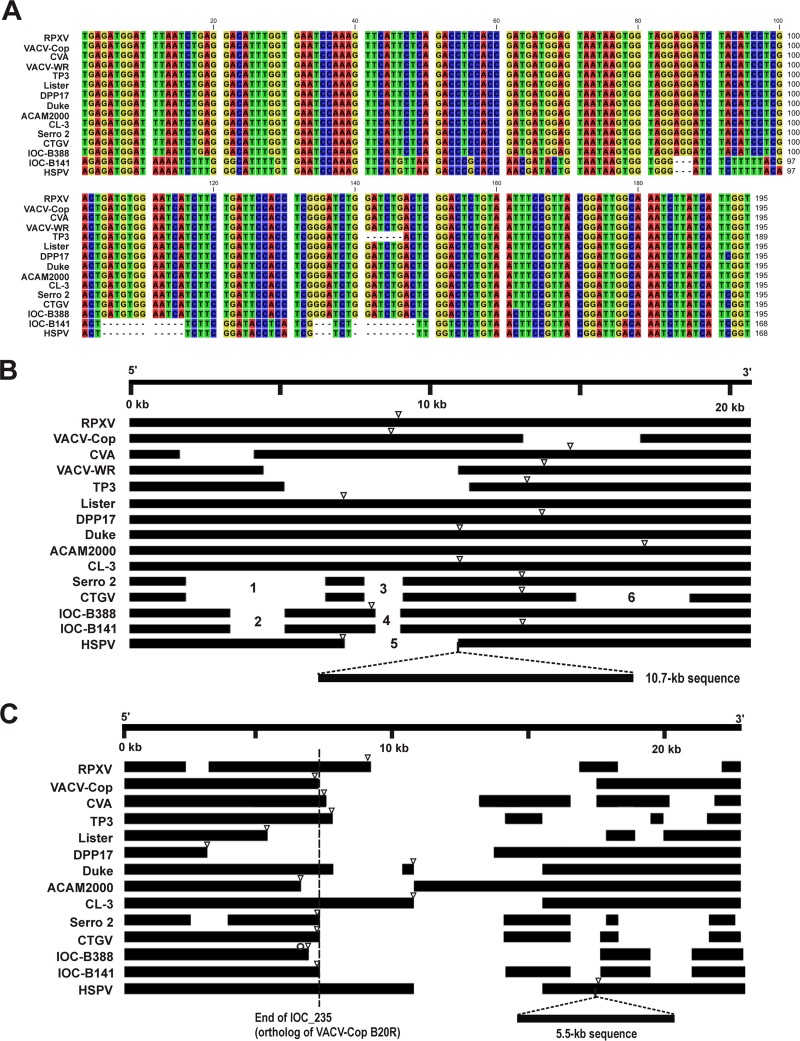
FIG 9.
Major differences in the genome terminal regions of B141, B388, other VACV strains, and HSPV. (A) Multialignment of a 168-nt region of orthologs of VACV-Cop C23L (IOC_003). Note that sequences of B141 and HSPV are identical, except for nt 97, and different from all other VACV strains, including B388. (B) Multialignment of the 5′ termini of different VACV genomes. Minor deletions and gaps were omitted in order to highlight major deletion patterns. Deletions marked as 1 to 6 are described in the text. Open triangles indicate the 5′ ITR junction sites in virus genomes. A 10.7-kb deletion, common to all VACV genomes, is represented as an insertion in the HSPV genome to facilitate genome comparison. (C) Scheme of ORF alignment of the 3′ termini based on a blast atlas. Note that, for the sake of clarity, the intergenic regions were removed as well as minor gaps and deletions. Open triangles indicate the 3′ ITR junction sites in virus genomes. The 5.5-kb deletion, common to all VACV, is represented as an insertion in the HSPV genome to facilitate genome comparisons. The dashed line indicates the end of gene IOC_235 (ortholog of VACV-Cop B20R). The open circle indicates the gene IOC_234 (ortholog of VACV-Cop B19R).