Abstract
Bordetella bronchiseptica can use catecholamines to obtain iron from transferrin and lactoferrin via uptake pathways involving the BfrA, BfrD, and BfrE outer membrane receptor proteins, and although Bordetella pertussis has the bfrD and bfrE genes, the role of these genes in iron uptake has not been demonstrated. In this study, the bfrD and bfrE genes of B. pertussis were shown to be functional in B. bronchiseptica, but neither B. bronchiseptica bfrD nor bfrE imparted catecholamine utilization to B. pertussis. Gene fusion analyses found that expression of B. bronchiseptica bfrA was increased during iron starvation, as is common for iron receptor genes, but that expression of the bfrD and bfrE genes of both species was decreased during iron limitation. As shown previously for B. pertussis, bfrD expression in B. bronchiseptica was also dependent on the BvgAS virulence regulatory system; however, in contrast to the case in B. pertussis, the known modulators nicotinic acid and sulfate, which silence Bvg-activated genes, did not silence expression of bfrD in B. bronchiseptica. Further studies using a B. bronchiseptica bvgAS mutant expressing the B. pertussis bvgAS genes revealed that the interspecies differences in bfrD modulation are partly due to BvgAS differences. Mouse respiratory infection experiments determined that catecholamine utilization contributes to the in vivo fitness of B. bronchiseptica and B. pertussis. Additional evidence of the in vivo importance of the B. pertussis receptors was obtained from serologic studies demonstrating pertussis patient serum reactivity with the B. pertussis BfrD and BfrE proteins.
INTRODUCTION
Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica are Gram-negative bacterial respiratory pathogens that have been termed the classical species of Bordetella. B. pertussis is a strict human pathogen that causes whooping cough (pertussis), which has been increasing in frequency (1–4). B. parapertussis causes a pertussis-like infection in humans and can also infect sheep, whereas B. bronchiseptica infects a range of nonhuman mammalian hosts as well as immunocompromised humans. The classical species of Bordetella are obligate pathogens that colonize the ciliated respiratory epithelium of the host, where they replicate and produce multiple virulence factors that promote their successful in vivo growth and cause symptoms of disease. Many genes, including most of the known Bordetella virulence factor genes, are positively regulated at the transcriptional level by the BvgAS two-component regulatory system (4–6).
Like virtually all other microbial pathogens, Bordetella species require iron for growth (7–9). Gram-negative bacteria generally rely on the TonB system to supply the energy for receptor-mediated transport of various iron sources across the bacterial outer membrane to the periplasm (10). B. bronchiseptica has 19 genes encoding known or predicted TonB-dependent outer membrane receptors, B. pertussis has 15 such genes, and B. parapertussis has 14 of the receptor genes (11, 12). In response to iron starvation, the classical bordetellae, including B. pertussis and B. bronchiseptica, produce and use the alcaligin siderophore for iron retrieval (13, 14) and can also use xenosiderophores, including enterobactin, produced by members of the Enterobacteriaceae (15); the fungal siderophore ferrichrome; and desferrioxamine B, produced by Streptomyces spp. (16). Bordetella species also use heme iron sources (17–19). Recently, we found that B. bronchiseptica can use neuroendocrine catecholamine hormones, such as norepinephrine, to obtain iron from the host glycoproteins transferrin and lactoferrin (20). The TonB-dependent outer membrane receptor proteins BfrA, BfrD, and BfrE were each determined to be involved in catecholamine-mediated utilization of transferrin iron. Each of these receptors also functioned in utilization of transferrin iron via both enterobactin and its breakdown product, 2,3-dihydroxybenzoylserine, in B. bronchiseptica.
Although the classical Bordetella species are highly genetically related, B. pertussis and B. parapertussis do not have the bfrA catecholamine receptor gene. B. pertussis has apparently intact bfrD and bfrE genes; however, its ability to use catecholamines to obtain iron from either transferrin or lactoferrin has not yet been demonstrated successfully (20). B. parapertussis has bfrD and an apparent bfrE pseudogene; the ability of this species to use norepinephrine to obtain iron from transferrin was modest, and it was unable to use norepinephrine to obtain iron from lactoferrin. The bfrD and bfrE genes lie adjacent to each other on the Bordetella chromosome, and bfrD appears to have arisen by duplication of bfrE (20). The bfrD and bfrE nucleotide sequences are 56.7% similar (21), and the deduced proteins have 57.5% pairwise identity. Although its involvement in iron acquisition was unknown at the time, BfrD was first identified in a study of outer membrane proteins that were produced in wild-type Bvg+ virulent-phase B. pertussis but were not produced by a bvgS mutant strain (22). A B. pertussis mutational analysis later showed that bfrD was transcriptionally activated by BvgAS and that its expression was decreased under modulating in vitro growth conditions that used nicotinic acid or sulfate to silence expression of Bvg-activated genes (21). Since B. pertussis BfrD was recently proposed as a potential pertussis vaccine candidate (23), more information about its function in B. pertussis would be useful.
In this characterization of the Bordetella catecholamine receptors, our analyses revealed that in these organisms, bfrA and bfrE transcriptional activities were modest, and bfrD expression appeared to be elevated under iron-replete growth conditions. In B. bronchiseptica, bfrD transcription was activated by Bvg but was relatively insensitive to modulation compared to the case in B. pertussis. We investigated the functionality of the B. pertussis BfrD and BfrE receptor proteins and demonstrated the involvement of catecholamine receptors in Bordetella growth during host respiratory infection.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Bordetella strains used in this study are listed in Table 1. Bordetella strains were cultured on Bordet-Gengou (BG) agar (1), and Escherichia coli strains were grown using Luria-Bertani broth or agar. Stainer-Scholte (SS) defined medium (24, 25) was used for Bordetella liquid cultures, and B. pertussis SS cultures were supplemented with 0.1 or 0.5% Casamino Acids. Glassware was acid cleaned and rinsed in distilled deionized water prior to use. Iron-replete SS medium contained 36 μM ferrous sulfate, and iron-depleted SS medium lacked an iron supplement and was deferrated using Chelex 100 resin (Bio-Rad Laboratories, Hercules, CA). In some studies, modulation conditions were achieved by addition of nicotinic acid or magnesium sulfate to SS medium at the indicated concentrations. Antibiotics were used as appropriate, at the following concentrations: tetracycline, 15 μg/ml; ampicillin, 100 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; and streptomycin, 20 μg/ml. Growth was monitored spectrophotometrically at a wavelength of 600 nm.
TABLE 1.
Bordetella strains used in this study
| Strain | Relevant genotype or description | Source or reference(s) |
|---|---|---|
| B. pertussis strains | ||
| BP338 | Tohama I derivative, clinical isolate, ca. 1954 | 5, 65 |
| BP347 | BP338; bvgS transposon mutant | 5 |
| UT25Sm1 | UT25; streptomycin resistant; clinical isolate, 1975 | 66, 67 |
| PM29 | UT25Sm1 with integrated chromosomal bfrD-lacZ fusion | This study |
| PM30 | UT25Sm1 with integrated chromosomal bfrE-lacZ fusion | This study |
| PM21 | UT25Sm1 with in-frame deletion in bfrD | This study |
| PM22 | UT25Sm1 with deletion of bfrD and bfrE | This study |
| B. bronchiseptica strains | ||
| RB50 | Rabbit isolate; streptomycin resistant | 68 |
| RB52 | RB50 bvgAS deletion mutant derivative containing bvgAS from B. pertussis BP338 | 52 |
| RB54 | RB50 bvgS deletion mutant derivative | 68 |
| RBB20 | RB50 bfrA bfrDE triple mutant; in-frame deletion in bfrA and deletion in bfrD-bfrE | This study |
| B013N | Swine isolate; nalidixic acid resistant | 69, 70 |
| BRM26 | B013N derivative; ΔalcA; alcaligin deficient | 71 |
General genetic procedures.
Standard methods were used for construction of recombinant plasmids (26). Reaction mixtures for PCR contained 5% dimethyl sulfoxide. Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA). Plasmids were transferred to E. coli strains by electroporation and to Bordetella strains by conjugation using E. coli DH5α as the donor strain, with plasmid pRK2013 providing mobilization functions (27). Plasmid pRK415 (28) was used as a cloning vector for Bordetella strains. pGEM3Z (Promega, Madison, WI) and pBlueScript II KS(+) (Agilent Technologies, Santa Clara, CA) were used as cloning vectors for E. coli.
The Bordetella genome sequences (http://www.ncbi.nlm.nih.gov/nucleotide) (B. pertussis Tohama I, accession no. NC_002929; B. parapertussis, accession no. NC_002928; and B. bronchiseptica RB50, accession no. NC_002927) were accessed at the GeneDB website (http://www.genedb.org/), developed and maintained by the Sanger Institute's Pathogen Sequencing Unit. DNA and protein analyses and PCR primer design used the Geneious, version 6.1.8, software package (Biomatters Ltd., Auckland, New Zealand).
Cloning of Bordetella receptor genes.
The Bordetella bfrD and bfrE genes, along with their native upstream sequences, were cloned into pRK415. bfrD was PCR amplified from the B. pertussis UT25 genomic DNA template by using primers bfrD-up (5′-CCCCAAGCTTAGCGCCAATCTGCCGTTTCTCA-3′) and bfrD-low (5′-CCCCTGCAGTATCCAGCAAGGGCGCCGTCATCT-3′). The 3.2-kb product was cloned into pBlueScript II KS(+) as a HindIII-PstI fragment, yielding plasmid pBKS+20, and then subcloned into pRK415, producing plasmid pRK71. Similarly, B. pertussis bfrE was PCR amplified using primers bfrE-up (5′-CCCCAAGCTTCAACGCAGATGCCGGCTACAACAA-3′) and bfrE-low (5′-CCCCTCTAGACCCGGGTGGCTCGATGGCAAACA-3′), cloned as a 2.8-kb HindIII-XbaI fragment into pBlueScript II KS(+), and subcloned into pRK415 (pRK72). B. bronchiseptica bfrD was PCR amplified using primers bfrD-up and bfrD-low, and the 3.2-kb product was cloned into pGEM3Z as a HindIII-PstI fragment and then subcloned into pRK415, resulting in plasmid pRK66. Construction of plasmid pRK67, containing B. bronchiseptica bfrE, was achieved by PCR amplification using primers bfrE-up and bfrE-low, cloning of the 2.8-kb product into pGEM3Z as a HindIII-XbaI fragment, and subcloning into pRK415.
Construction and analysis of transcriptional lacZ fusions.
Low-copy-number plasmid-borne B. bronchiseptica bfrA, bfrD, and bfrE promoter fusions to lacZ were constructed using the gene fusion plasmid vector pMP220 (29). A 3.3-kb DNA fragment encompassing the predicted promoter region of bfrA was amplified by PCR, using primers bfrA-A (5′-GGCCAAGCTTGTGCGCACGACCGGTTTTGACG-3′) and bfrA-B (5′-GGCCAAGCTTGCAGCCCGCGGTGGTGGAGAAG-3′), and cloned into pBlueScript II KS(+) by using the primer HindIII adapter sites. A 1.1-kb BamHI-PstI DNA subfragment was cloned into a pGEM3Z intermediate and then excised with KpnI-PstI and subcloned into pMP220 to produce the bfrA-lacZ fusion plasmid pMP6. Primers bfrD-up and bfrD-low were used to PCR amplify a 3.2-kb product containing the B. bronchiseptica RB50 bfrD region. Digestion of the 3.2-kb product with HindIII and EcoRI yielded a 930-bp DNA fragment that was cloned (pBKS+15) and subsequently subcloned into pMP220 as a KpnI-PstI fragment to produce pMP7. The B. bronchiseptica bfrE promoter region was cloned into pMP220 as a 238-bp PCR product (amplified with primers PbfrE3 [5′-CCCGAATTCATGCTGACGTTCAAGCTGAG-3′] and PbfrE4 [5′-CCCCTGCAGAAAAATCTGAATAACAACGATTC-3′]), using EcoRI and PstI primer adapters, to yield plasmid pMP8. A cyaA-lacZ gene fusion plasmid was constructed by PCR amplification of the B. pertussis UT25 cyaA promoter region (30), using primers cyaP3 (5′-GGCCGAATTCGAGTTCGGTGTCGGCGTCCATTAG-3′) and cyaP4 (5′-GGCCCTGCAGTGTGTAGCGCTCAGAACCTCATCC-3′), cloning of the 624-bp PstI DNA subfragment into pGEM3Z, and excision of the insert DNA fragment with KpnI and SphI for subcloning into pMP220 to produce pMP2. Fusion plasmids were transferred to B. bronchiseptica and B. pertussis strains by triparental mating.
Single-copy chromosomal lacZ fusion construction used the integrative fusion plasmid pFUS2 (21). A 351-bp PCR product consisting of the bfrD promoter region of B. pertussis strain UT25 was cloned into pFUS2 by using BamHI and KpnI adapters (5′-CCGGTACCGCCACCGTCAAATGGGA-3′ and 5′-CCGGATCCCAGGGCGGGATTGTTGCCGT-3′); similarly, a 402-bp B. pertussis bfrE fragment was cloned by using BamHI and KpnI adapters (5′-CCGGTACCCGGAGAAAAGCCGCACCGT-3′ and 5′-CCGGATCCATTGCGACCGTTGTCCGCGT-3′). Each pFUS2 construct was conjugally transferred to B. pertussis strain UT25Sm1, and plasmid integrants at bfrD and bfrE were selected on BG agar containing gentamicin, streptomycin, and colicin B (31).
Transcriptional activities of lacZ fusion genes were monitored using β-galactosidase assays (32). Bordetella strains carrying gene fusion plasmids or chromosomal gene fusions were grown in iron-replete SS medium with antibiotic selection and then subcultured in iron-replete SS medium, iron-depleted SS medium, or iron-replete SS medium containing supplements, as indicated. After 24 and 48 h of growth, LacZ activities were measured. Values are reported as mean LacZ activities (in Miller units; n = 3) ± 1 standard deviation (SD). The results presented are representative of at least two experimental trials. Mean LacZ activities were compared using Student's paired t test (two-tailed distribution, hypothesized difference = 0) in StatView, version 4.51, software (Abacus Concepts, Inc.). The P value is the probability that the mean LacZ activity is the same between strains or under different growth conditions, and P values of ≤0.05 were considered to be significant.
Construction of Bordetella mutant strains.
A bfrA bfrD bfrE triple mutant was constructed in the B. bronchiseptica RB50 strain background. Briefly, a bfrD bfrE deletion mutant was first constructed as described previously (20). bfrD and bfrE are adjacent to each other on the chromosome, and the mutation resulted in the removal of a 4,393-bp DNA segment spanning the genes. The bfrDE deletion was transferred to strain RB50 by allelic exchange to produce strain RBB19, and the deletion was confirmed by PCR. RBB19 was then made bfrA by using the allelic exchange plasmid pEG7.31, carrying a bfrA deletion mutation (20). The 810-bp in-frame bfrA deletion mutation was confirmed by PCR and yielded the ΔbfrA ΔbfrDE triple mutant RBB20. For construction of a B. pertussis bfrD mutant, pBKS+20 was cut with AatII and religated, resulting in a 1,392-bp in-frame deletion in bfrD. The ΔbfrD allele was subcloned into the suicide plasmid pSS1129 (33) as a BamHI-HindIII fragment, yielding pSS39. The mutation was transferred to B. pertussis UT25Sm1 by allelic exchange and confirmed by PCR.
Growth assays.
Norepinephrine-mediated growth stimulation by transferrin was measured by determining the optical density at 600 nm (OD600) of liquid SS cultures as described previously (20). B. bronchiseptica strains were grown on BG agar for 24 h, subcultured into iron-replete SS medium, and grown for 18 h. The cells were washed with SS basal medium lacking iron and used as inocula for the test cultures, at an initial OD600 of 0.01. Test media included iron-replete SS medium or iron-depleted SS medium containing 10 mM sodium bicarbonate and 200 μg/ml transferrin (approximately 2.5 μM; 30% iron saturated), with or without 50 μM norepinephrine. For B. pertussis strains, growth from BG agar was used to inoculate iron-replete SS medium. After growth for 24 h, the bacteria were harvested, washed, and subcultured into test media as described for B. bronchiseptica growth assays, but at an initial OD600 of 0.02. Growth yield results are reported as the means of results from triplicate cultures, and each experiment was performed at least twice independently. Mean growth yields were compared using Student's paired t test (two-tailed distribution, hypothesized difference = 0) in StatView, version 4.51, software (Abacus Concepts, Inc.). The P value is the probability that the mean growth yield is the same between strains or growth conditions, and P values of ≤0.05 were considered to be significant.
Mouse infections.
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of Minnesota, and all procedures conformed to the guidelines of the Guide for the Care and Use of Laboratory Animals (34). Bordetella inocula for mouse competition infection experiments were prepared as described previously (35, 36). Wild-type B. bronchiseptica strain RB50 and the isogenic ΔbfrA ΔbfrDE triple mutant strain RBB20 were cultivated on BG agar and then subcultured onto iron-replete SS medium and grown for 24 h. The bacteria were washed and resuspended in 0.9% NaCl, and the two strain suspensions were combined at an estimated 1:1 strain ratio to prepare a mixed-strain suspension determined by CFU counting to have 9.3 × 105 total CFU/ml. Wild-type B. pertussis strain UT25Sm1 and the isogenic ΔbfrD mutant strain PM21 were grown on BG agar, subcultured onto SS medium containing 0.5% Casamino Acids, and then combined to produce a 1:1 strain ratio inoculum containing 2.5 × 107 total CFU/ml. BALB/cAnNHsd mice (10 to 20 g) (Harlan Sprague Dawley, Inc.) were briefly sedated by isoflurane inhalation and then infected intranasally with a 20-μl volume containing ∼5 × 105 total CFU of a 1:1 mixture of B. bronchiseptica strains RB50 and RBB20 or, for B. pertussis, 1 × 106 total CFU of UT25Sm1 and PM21.
At the indicated time points postinfection, 5 mice were euthanized, and lungs and tracheal homogenates from individual mice were plated on BG agar for total CFU determination. The nasal tissue homogenates from each mouse group of 5 were pooled and plated. Differential enumeration of the bacterial strains was accomplished by picking 100 colonies, patching them onto BG agar, and performing in situ DNA hybridizations at high stringency, as described previously (26). The 0.9-kb bfrD-specific DNA hybridization probe was PCR amplified using primers bfrD1 and bfrD2 and Bordetella genomic DNA templates and then radiolabeled with [α-32P]dCTP (MP Biomedicals, Santa Ana, CA) by using the Random Primers DNA labeling system (Invitrogen). The competitive index (CI) was calculated as the mutant/wild-type CFU ratio in the output recovered at each time point divided by the mutant/wild-type CFU ratio in the input inoculum. Three experimental trials were conducted. Student's t test was used to determine whether the mean CI at each time point differed significantly from the hypothesized mean value of 1.00 (the predicted mean CI if there were no difference in fitness between the two strains) and from the mean CI at the preceding time point. P values of ≤0.05 were considered to be significant.
Analysis of Bordetella proteins.
Recombinant BfrD and BfrE proteins were produced in E. coli. The bfrD coding sequence minus the predicted signal sequence was PCR amplified from B. pertussis strain UT25 by using primers His-bfrD up (5′-GGGCTCGAGGCGTCCACGGCGGTCCAG-3′) and His-bfrD low (5′-GGGGAAGCTTGGGACGCTTCGCCCTCTA-3′). The 2.5-kb PCR product was cloned into the expression vector pBAD/His-A (Invitrogen) as an XhoI-HindIII DNA fragment (pBAD/bfrD). Similarly, the bfrE sequence minus the predicted signal sequence was amplified from strain UT25 by using primers His-bfrE up (5′-GGGCTCGAGACCAGCGCGGGCGTTACC-3′) and His-bfrE low (5′-GGGGAAGCTTGCAATGCGCCGGTTTTCC-3′). The 2.2-kb PCR product was cloned into pBAD/His-A as an XhoI-HindIII fragment (pBAD/bfrE). Plasmids were transformed into E. coli strain TOP10 (Invitrogen) via electroporation. A 25-ml Luria-Bertani broth culture containing 200 μg/ml ampicillin and 0.02% arabinose inducer was inoculated to an initial OD600 of 0.04 with TOP10(pBAD/bfrD) or TOP10(pBAD/bfrE) cells taken directly from the primary transformation plates. The cultures were incubated at 35°C for 4 h, and the cells were harvested and suspended to 20 OD600 units/ml in 50 mM HEPES, pH 7.4.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5% polyacrylamide gels, and proteins were visualized by staining with Coomassie blue dye. For immunoblot studies, bacterial sample suspensions were normalized by using the OD600 and heated to 100°C in solubilization buffer, and the cleared lysate was loaded onto 7.5% polyacrylamide gels for electrophoresis. Purified BfrD and BfrE proteins were obtained by cutting relevant protein bands from electrophoretic gels. BfrD and BfrE protein identities were confirmed by tandem mass spectrometry (MS/MS), and proteins in the remainder of the gel slices were subjected to electrophoresis as indicated.
Research involving materials from human subjects was performed in accordance with federal guidelines and institutional policies. Immunoblot analyses used sera from B. pertussis culture-positive patient donors (aged 6 to 17 years) who had previously been vaccinated and sera from a control group of healthy adult donors not known to have had previous B. pertussis infection. Serum samples were provided by Alison Weiss, University of Cincinnati (37), or were from our laboratory collection. The sera were analyzed for antibody reactivity with the recombinant B. pertussis BfrD and BfrE proteins by using standard immunoblotting methods, as described previously (38). Blots with control and patient sera were performed in parallel on replicate gels, using the same serum dilution. The blots were developed using the same developer solution and were imaged together on a flatbed scanner, using identical image settings.
Reverse-phase LC-MS/MS analysis.
The putative BfrD and BfrE protein bands were excised from Coomassie blue-stained SDS-PAGE gels and treated with trypsin. Trypsin treatment and liquid chromatography-MS/MS (LC-MS/MS) were performed at the University of Minnesota Center for Mass Spectrometry and Proteomics facility. Peptide products were separated on a C18 Nanotrap column (Michrom BioResources, Auburn, CA) and subjected to tandem mass spectrometry using a ThermoFinnigan (ABI, Inc., Foster City, CA) LTQ ion-trap mass spectrometer (MS). Mass spectrometry data were analyzed using Sequest (version 27, rev. 12; Thermo Fisher Scientific, San Jose, CA). Sequest was set up to search the NCBI protein sequence database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein), including E. coli proteins and the B. pertussis Tohama I BfrD (accession no. NP_879666.1) and BfrE (accession no. NP_879667.1) proteins, assuming trypsin cleavage. Sequest was searched with a fragment ion mass tolerance of 1.00 Da. An iodoacetamide derivative of cysteine was specified as a fixed modification, and oxidation of methionine was set as a variable modification. Scaffold (version Scaffold_4.4.1; Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >95.0% probability as specified by the Peptide Prophet algorithm (39). Protein identifications were accepted if they could be established at >99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (40). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
RESULTS
The B. pertussis BfrD and BfrE receptors are functional.
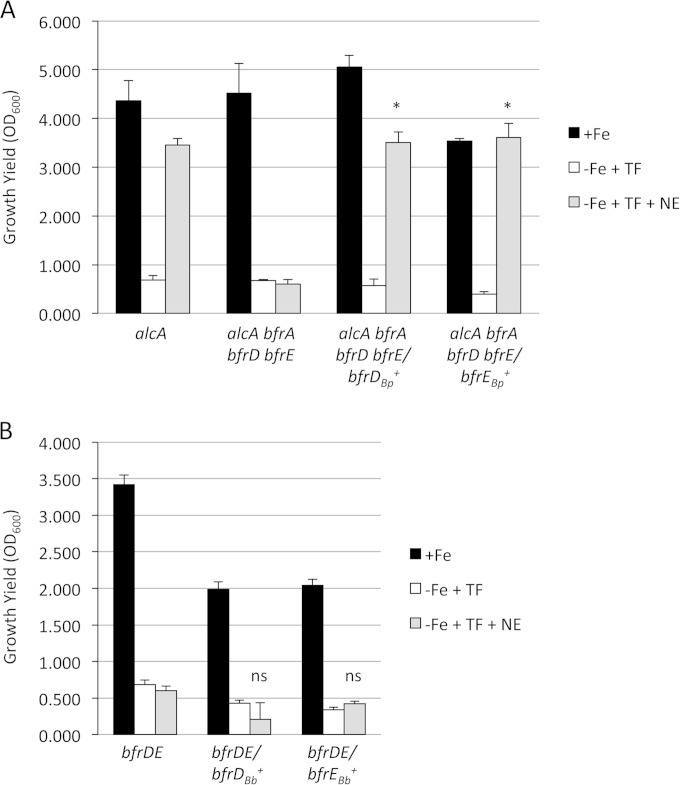
In B. bronchiseptica, the BfrA, BfrD, and BfrE receptors each promote catecholamine-mediated uptake of transferrin iron. B. pertussis has the bfrD and bfrE genes (99.9% and 99.8% identical to those of B. bronchiseptica, respectively), but we have not yet been able to demonstrate the utilization of catecholamines for iron acquisition by B. pertussis (20). Since it is possible that the B. pertussis BfrD and BfrE proteins are not functional, the B. pertussis genes were cloned and transferred to B. bronchiseptica strain RBB21, a bfrA bfrD bfrE mutant that is unable to use catecholamines to obtain transferrin iron and also lacks alcaligin production (alcA). The plasmid-bearing strains were examined for the ability to grow in iron-replete SS defined medium and iron-depleted SS medium supplemented with 30% iron-saturated transferrin as the sole iron source, either lacking or containing 50 μM norepinephrine (Fig. 1A). As expected, compared with the RBB22 alcA parent strain, the bfrA bfrD bfrE mutant derivative RBB21 grew poorly with norepinephrine as the only means to obtain transferrin iron. However, RBB21 carrying the B. pertussis bfrD or bfrE gene grew as well as the RBB22 parent strain in norepinephrine-supplemented medium. These results indicate that B. pertussis bfrD and bfrE encode receptors that function in norepinephrine-mediated iron retrieval in B. bronchiseptica.
FIG 1.
The B. pertussis bfrD and bfrE genes confer a norepinephrine-mediated transferrin utilization phenotype to B. bronchiseptica, but the B. bronchiseptica bfrD and bfrE genes do not promote norepinephrine-mediated transferrin utilization in B. pertussis. (A) B. bronchiseptica strains RBB22 (alcA) and RBB21 (alcA bfrA bfrD bfrE), each carrying the pRK415 plasmid vector, and RBB21, carrying the B. pertussis bfrD+ plasmid pRK71 or bfrE+ plasmid pRK72, were cultured in iron-depleted SS medium with transferrin, either without norepinephrine (−Fe + TF) or with 50 μM norepinephrine (−Fe + TF + NE). Iron-replete control cultures contained 36 μM iron (+Fe). Growth yields (n = 3; values are means and SD) were determined after 48 h. (B) B. pertussis strain PM22 (bfrD bfrE) carrying the pRK415 plasmid vector and PM22 carrying the B. bronchiseptica bfrD+ (pRK66) or bfrE+ (pRK67) plasmid were grown in SS media as described for panel A. Growth yields (n = 3; means and SD) were determined after 48 h. Statistically significant differences in mean growth yields in −Fe + TF + NE cultures compared with control strains were calculated using Student's paired t test as described in Materials and Methods. *, P ≤ 0.05; ns, not significant.
The B. bronchiseptica bfrD and bfrE genes were tested for the ability to confer norepinephrine utilization on B. pertussis. The growth of the B. pertussis bfrD bfrE mutant PM22 carrying the plasmid-borne B. bronchiseptica bfrD or bfrE gene was not stimulated by norepinephrine in the presence of transferrin (Fig. 1B). The identities of the presumptive B. bronchiseptica cytoplasmic membrane transporter genes required for catecholamine utilization are not yet known. The lack of observed in vitro B. pertussis catecholamine utilization is not likely due to nonfunctional BfrD or BfrE receptor proteins but may be due to an absent or defective cytoplasmic membrane transport apparatus or other catecholamine utilization function, or it may be a technical limitation of the in vitro growth assay system used for B. pertussis.
Analyses of bfrA, bfrD, and bfrE transcription.
Bacterial iron transport genes are typically repressed by Fur (or functionally similar proteins) with its bound iron corepressor under iron-replete growth conditions. When internal iron stores are depleted, Fur is inactive and the iron assimilation genes are derepressed (41, 42). In addition, some iron system genes are positively regulated by mechanisms involving the cognate iron source acting as the inducer (9, 43). In a microarray study of the transcriptional responses of B. pertussis and B. bronchiseptica to iron starvation stress, the bfrA, bfrD, and bfrE genes were not identified as being significantly iron regulated (44). Using the consensus Fur binding sequence 5′-GATAATGATAATCATTATC-3′ (45, 46) as the query, in silico analysis of the bfrA, bfrD, and bfrE upstream DNA regions identified potential Fur binding sites. Three overlapping sites, with 7 or 8 mismatches, are positioned 46, 52, and 58 nucleotides (nt) upstream of bfrA. As reported by Passerini de Rossi et al. (47), one potential 19-nt Fur binding site (8 mismatches) is located 121 nt upstream of bfrD. bfrD and bfrE are adjacent on the chromosome but are separated by 211 bp and are transcribed independently (20). A possible Fur binding site with 8 mismatches is positioned 12 bp upstream of the bfrE coding sequence. The ability of the upstream regions of these genes to bind Fur in vivo was examined using the Fur titration assay (48). In this E. coli assay, a functional Fur binding site introduced on a multicopy plasmid can titrate Fur, thus relieving the repression of a chromosomal fhuF-lacZ fusion that is normally repressed under iron-replete growth conditions, resulting in a LacZ+ phenotype. Compared with plasmid vector controls and a plasmid bearing the consensus Fur binding sequence which demonstrated strong Fur binding (LacZ) activity, the upstream DNA regions of bfrA, bfrD, and bfrE exhibited variable, weak Fur binding (data not shown). To evaluate the transcriptional regulation of the catecholamine utilization genes, gene fusions were analyzed in Bordetella cells.
bfrA.
Of the three classical Bordetella species, only B. bronchiseptica has the bfrA receptor gene. B. bronchiseptica wild-type strain RB50 carrying a low-copy-number bfrA-lacZ transcriptional fusion plasmid (pMP6) was grown in iron-replete SS defined medium or iron-restricted SS medium containing transferrin (30% iron saturated) as the sole iron source (Fig. 2). Bordetella bfrA transcription levels were low in iron-replete medium but were elevated upon iron restriction in the presence of transferrin; the presence or absence of norepinephrine did not influence bfrA expression. Compared with our previous siderophore and heme receptor gene-lacZ fusion analyses of iron-replete or iron-starved Bordetella cells (49–51), the overall bfrA expression levels were quite low. Similar results were obtained using other B. bronchiseptica host strains (data not shown). LacZ transcriptional fusions provide an established, reliable measure of Bordetella promoter activity, and the growth media and conditions that were used do not produce artifactual LacZ assay results. Similarly low bfrA expression levels were observed in our transcriptome studies of iron-replete versus iron-starved B. bronchiseptica strain RB50 (44). Experiments using iron-replete or iron-restricted SS medium that lacked transferrin showed comparable B. bronchiseptica bfrA transcription levels, indicating that it was the lack of iron, not the presence of transferrin, that promoted bfrA gene expression (data not shown).
FIG 2.
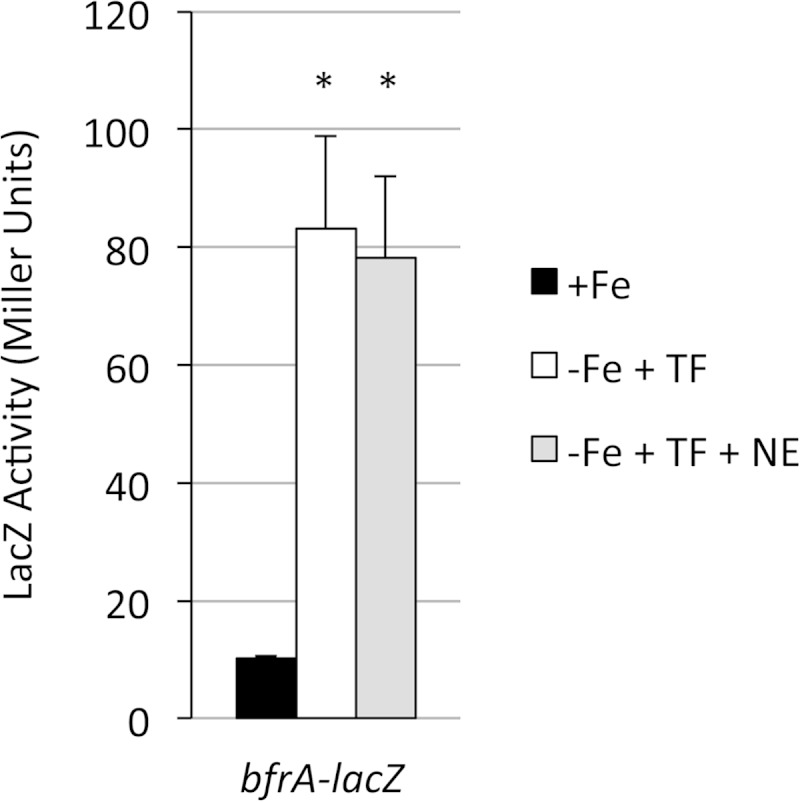
Transcriptional regulation of B. bronchiseptica bfrA. RB50(pMP6) (bfrA-lacZ) was grown in iron-replete (+Fe) SS medium or in iron-depleted medium with transferrin in the absence (−Fe + TF) or presence (−Fe + TF + NE) of norepinephrine. LacZ activity (n = 3; means and SD) was measured after 24 h of growth. *, P ≤ 0.05 for comparison to +Fe cultures. There was no significant difference in bfrA-lacZ expression in cells grown in −Fe + TF medium and those grown in −Fe + TF + NE medium.
bfrE.
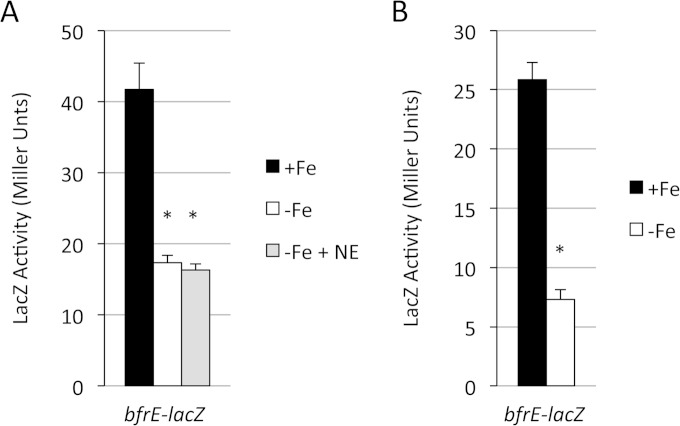
In B. bronchiseptica strain RB50, bfrE-lacZ expression (pMP8) was low under all SS medium growth conditions (Fig. 3A). There was no effect of norepinephrine on bfrE-lacZ expression. The presence of iron in the medium was associated with elevated bfrE transcriptional activity compared with that under iron-restricted conditions. As observed for bfrA, addition of transferrin did not alter transcription of bfrE (data not shown). A different B. bronchiseptica host strain showed similar bfrE-lacZ expression patterns (data not shown). To assess bfrE expression in B. pertussis, the promoter of the B. pertussis bfrE gene was fused to lacZ and integrated into the chromosome of strain UT25Sm1 at the bfrE locus. In this human-adapted species, transcription of bfrE was quite modest, and as with B. bronchiseptica, it was elevated under iron-replete conditions (Fig. 3B); expression was not affected by the presence of norepinephrine (data not shown). These results indicate that in B. bronchiseptica and B. pertussis grown under these culture conditions, expression levels of bfrE are generally low but are elevated under iron-replete conditions.
FIG 3.
Transcriptional regulation of B. bronchiseptica and B. pertussis bfrE. (A) B. bronchiseptica strain RB50(pMP8) (bfrE-lacZ) was cultured in iron-replete (+Fe) SS medium or in iron-depleted medium in the absence (−Fe) or presence (−Fe + NE) of norepinephrine. LacZ activity (n = 3; means and SD) was measured after 24 h of growth. (B) B. pertussis strain PM30, with the lacZ fusion plasmid pFUS2 integrated at the chromosomal bfrE locus, was grown under iron-replete (+Fe) or iron-depleted (−Fe) conditions, and LacZ activity (n = 3; means and SD) was measured after 48 h of growth. *, P ≤ 0.05 for comparison to +Fe cultures. For B. bronchiseptica RB50 in panel A, there was no significant difference in bfrE-lacZ expression between cells grown in −Fe versus −Fe + NE medium.
bfrD.
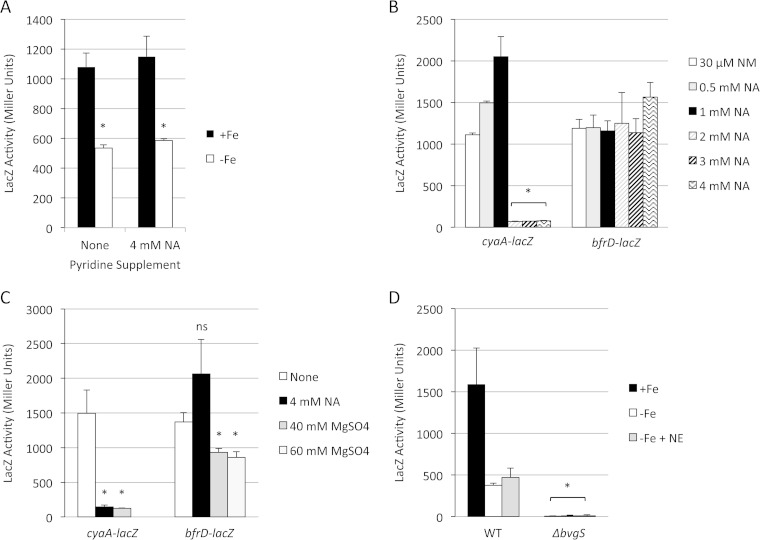
Transcription of bfrD in B. bronchiseptica was monitored in strain RB50 carrying the low-copy-number bfrD-lacZ transcriptional fusion plasmid pMP7. Similar to the pattern observed with bfrE, bfrD expression levels in RB50 were high under iron-replete conditions and decreased when bacteria were grown under iron-depleted conditions (Fig. 4A), but overall bfrD expression levels were much higher than those of bfrE. Iron titration experiments showed optimal bfrD expression when cells were cultured at iron concentrations ranging from 4.5 μM to 36 μM; growth with iron concentrations of 1 to 2 μM resulted in significantly lower bfrD-lacZ expression levels (data not shown).
FIG 4.
Effects of iron starvation and the BvgAS system on transcriptional regulation of B. bronchiseptica bfrD. (A) RB50(pMP7) was grown in iron-replete (+Fe) SS medium or iron-depleted medium (−Fe) in the absence or presence of the Bvg modulator nicotinic acid (NA; 4 mM), and LacZ activity was measured after 24 h of growth (n = 3; means and SD). *, P ≤ 0.05 for comparison to +Fe cultures. There was no statistically significant difference in bfrD-lacZ expression between cells grown with 4 mM NA and those grown in its absence. (B) RB50 carrying either pMP7 (bfrD-lacZ) or the known Bvg-responsive cyaA-lacZ control plasmid pMP2 was grown in iron-replete SS medium supplemented with either the standard pyridine supplement 30 μM nicotinamide (NM) or nicotinic acid (NA) at the indicated concentrations, and LacZ activity was measured after 24 h of growth (n = 3; means and SD). For cyaA-lacZ data, statistically significant differences for comparing cells grown in NM to those grown in 2 to 4 mM NA are indicated as follows: *, P ≤ 0.05. There were no statistically significant differences in bfrD-lacZ expression between cells grown in NM and those grown in NA at any concentration. (C) RB50(pMP2) (cyaA-lacZ) and RB50(pMP7) (bfrD-lacZ) were grown in standard SS medium (none) or in the same medium supplemented with nicotinic acid (NA) or magnesium sulfate at the indicated concentrations. LacZ activity was measured after 24 h of growth (n = 3; means and SD). Statistically significant differences in cells grown with the modulator NA or MgSO4 and those grown in the absence of modulators are indicated as follows: *, P ≤ 0.05 (ns, not significant). (D) RB50 (WT) and its ΔbvgS derivative RB54, each carrying the bfrD-lacZ fusion plasmid pMP7, were grown in iron-replete (+Fe) SS medium or in iron-depleted medium in the absence (−Fe) or presence (−Fe + NE) of norepinephrine. LacZ activity was measured after 24 h of growth (n = 3; means and SD). *, P ≤ 0.05 for comparing bfrD expression in strains RB54 and RB50 under each of the three growth conditions. There were no statistically significant differences between cells grown in −Fe and −Fe + NE media.
In B. pertussis, bfrD was reported to be a highly expressed Bvg-activated gene, exhibiting no significant expression in bvg mutant strains and decreased expression in the presence of modulator compounds, such as nicotinic acid or magnesium sulfate (21). As a first step toward determining whether bfrD expression is regulated by Bvg in B. bronchiseptica, wild-type strain RB50(pMP7) was grown in the presence or absence of modulating compounds, and bfrD expression was measured (Fig. 4A and B). As predicted, expression of the Bvg-activated, adenylate cyclase-encoding cyaA-lacZ fusion control was dramatically decreased when RB50(pMP2) was cultured in the presence of 2 to 4 mM nicotinic acid compared with expression levels when the strain was cultured in the absence of nicotinic acid (Fig. 4B). In contrast, nicotinic acid had no modulating effect on bfrD-lacZ expression in B. bronchiseptica (Fig. 4A and B). Compared with expression of cyaA, expression of bfrD was also significantly less sensitive to the modulating influence of sulfate (Fig. 4C); thus, bfrD expression is relatively insensitive to these modulators in B. bronchiseptica. As with bfrA and bfrE, norepinephrine supplementation of cultures had no effect on expression of bfrD (Fig. 4D). To determine whether bfrD transcription is Bvg dependent in B. bronchiseptica, as it is in B. pertussis, bfrD-lacZ expression was measured in wild-type RB50 compared with its isogenic ΔbvgS mutant derivative RB54 (Fig. 4D). Expression of bfrD in strain RB54 was negligible under all growth conditions. These results demonstrate that in B. bronchiseptica, transcription of bfrD is strictly Bvg dependent but is insensitive to nicotinic acid and sulfate modulation.
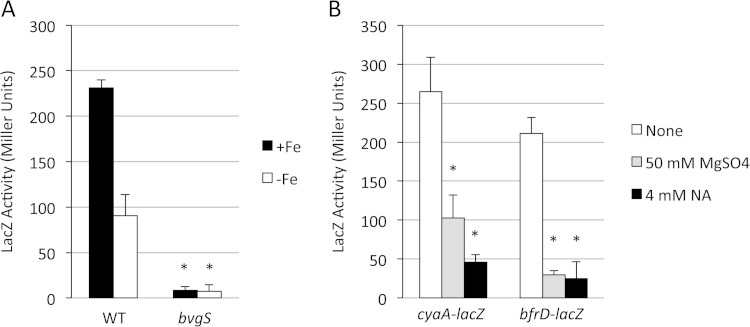
The same pMP7 plasmid, containing the B. bronchiseptica bfrD promoter region fused with lacZ, was transferred to B. pertussis wild-type strain BP338 and its bvgS mutant derivative BP347. Similar to the B. bronchiseptica results, transcription of bfrD in B. pertussis was greater under iron-replete than iron-restricted growth conditions, and expression was severely reduced in the bvgS mutant strain BP347 (Fig. 5A). However, in contrast to the case in B. bronchiseptica, bfrD expression in B. pertussis was modulated by sulfate and nicotinic acid, as was that of the cyaA-lacZ control (Fig. 5B). A single-copy B. pertussis bfrD-lacZ fusion was integrated into the chromosome of a different B. pertussis strain (UT25Sm1). As with the pMP7-borne B. bronchiseptica-derived bfrD-lacZ fusion, expression of the B. pertussis chromosomal bfrD-lacZ fusion was higher under iron-replete conditions (data not shown) and was subject to sulfate- and nicotinic acid-mediated modulation (Fig. 6).
FIG 5.
Transcription of B. bronchiseptica bfrD in B. pertussis is regulated by Bvg and modulated by nicotinic acid and sulfate. (A) Wild-type (WT) B. pertussis strain BP338 and the isogenic bvgS mutant strain BP347, both carrying pMP7 (bfrD-lacZ), were grown in iron-replete (+Fe) SS medium or iron-depleted medium (−Fe), and LacZ activity was measured after 48 h (n = 3; means and SD). *, P ≤ 0.05 for differences in bfrD-lacZ expression between BP338 and BP347 under each growth condition. For comparison of the LacZ activity of BP338(pMP7) in −Fe compared with +Fe medium, the P value was ≤0.05. (B) BP338 carrying either the cyaA-lacZ (pMP2) or bfrD-lacZ (pMP7) plasmid was grown in standard SS medium with no additions (none) or supplemented with the indicated concentrations of magnesium sulfate or nicotinic acid (NA) as a modulator. LacZ activity was measured after 24 h of growth (n = 3; means and SD). *, P ≤ 0.05 for comparing fusion gene expression in strain BP338 under nonmodulating (none) versus modulating growth conditions.
FIG 6.
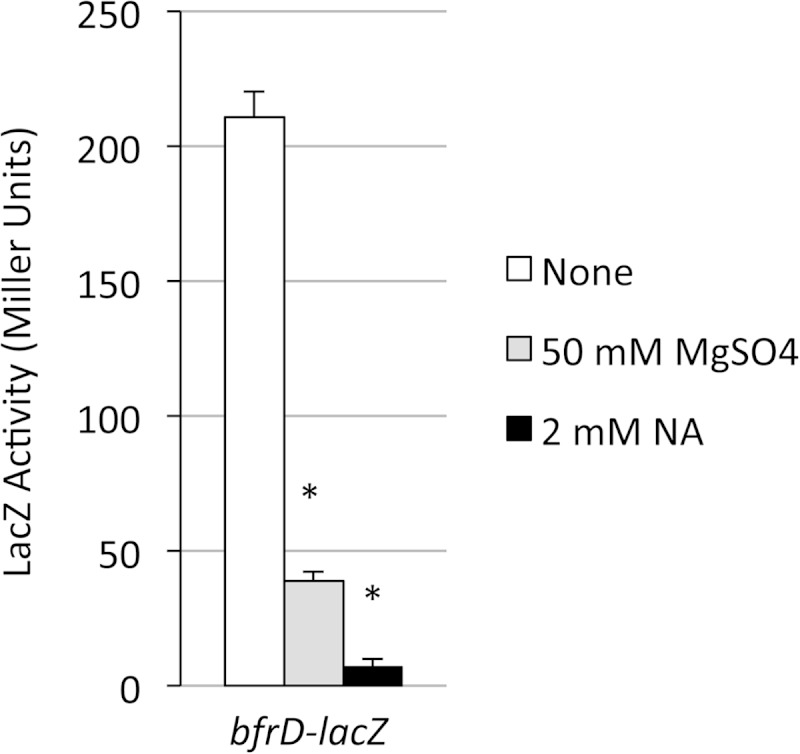
Transcription of the B. pertussis bfrD gene in B. pertussis is modulated by nicotinic acid and sulfate. B. pertussis strain PM29, with the lacZ fusion plasmid pFUS2 integrated at the chromosomal bfrD locus, was grown in standard SS medium with no additions (none) or with the indicated concentrations of magnesium sulfate or nicotinic acid (NA) as a modulator. LacZ activity was measured after 24 h of growth (n = 3; means and SD). *, P ≤ 0.05 for comparing bfrD expression in cells grown under modulating versus nonmodulating conditions.
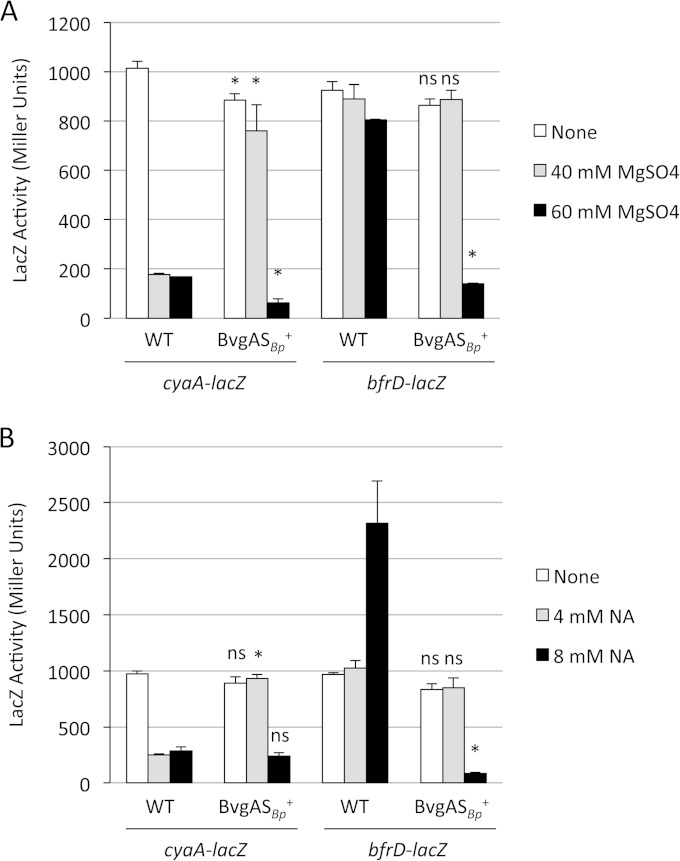
B. pertussis and B. bronchiseptica have been reported to vary in their sensitivity to modulator compounds, and differences in their BvgS proteins were implicated (52). In those studies, a derivative of B. bronchiseptica RB50, RB52, was constructed by replacing its bvgAS genes with those of B. pertussis strain BP338. In the present study, the bfrD-lacZ plasmid pMP7 was examined in strain RB52 for transcriptional responses to modulator compounds. Compared with the expression levels in the wild-type B. bronchiseptica RB50 host, in B. bronchiseptica RB52 the B. pertussis bvgAS alleles conferred increased sensitivity of bfrD-lacZ expression to modulation, with markedly decreased transcription at 60 mM MgSO4 (Fig. 7A). Conversely, in RB52, cyaA-lacZ expression showed a decreased sensitivity to sulfate modulation compared to the expression in RB50. Modulation using 4 mM and 8 mM nicotinic acid yielded bfrD and cyaA expression patterns similar to those seen with MgSO4 modulation in the two B. bronchiseptica strains (Fig. 7B). For unknown reasons, growth of the wild-type strain in 8 mM nicotinic acid consistently appeared to stimulate bfrD transcription. Overall, these results indicate that the relative resistance of bfrD transcription to modulation in B. bronchiseptica compared with that in B. pertussis is due in part to species-specific differences in BvgAS. However, since bfrD transcription in B. pertussis BP338 (the source of bvgAS in RB52) was effectively modulated by 4 mM nicotinic acid and 50 mM MgSO4 (Fig. 5B), other cellular differences are also likely to influence expression.
FIG 7.
Transcription of bfrD in B. bronchiseptica RB52, which has the B. bronchiseptica bvgAS genes replaced by the orthologous bvgAS genes of B. pertussis, is modulated by nicotinic acid and sulfate. Wild-type strain RB50 (WT) and its isogenic derivative RB52 (BvgASBp+), carrying pMP2 (cyaA-lacZ) or pMP7 (bfrD-lacZ), were grown in standard SS medium with no additions (none) or supplemented with the indicated concentrations of magnesium sulfate (A) or nicotinic acid (NA) (B) as a modulator. LacZ activity was measured after 24 h of growth (n = 3; means and SD). *, P ≤ 0.05 for comparing fusion gene expression in strains RB52 and RB50 grown under each condition; ns, not significant.
In aggregate, these results indicate that in B. pertussis and B. bronchiseptica, the transcriptional activity of bfrD is higher under iron-replete than iron-restricted growth conditions. Furthermore, in B. bronchiseptica, bfrD is under strict transcriptional control of the BvgAS system, as it is in B. pertussis. B. bronchiseptica transcription of bfrD was not modulated by compounds given at concentrations known to decrease expression of other Bvg-activated genes, as in B. pertussis, despite strong interspecies conservation of the putative bfrD upstream control regions.
Contributions of BfrA, BfrD, and BfrE to Bordetella growth in vivo.
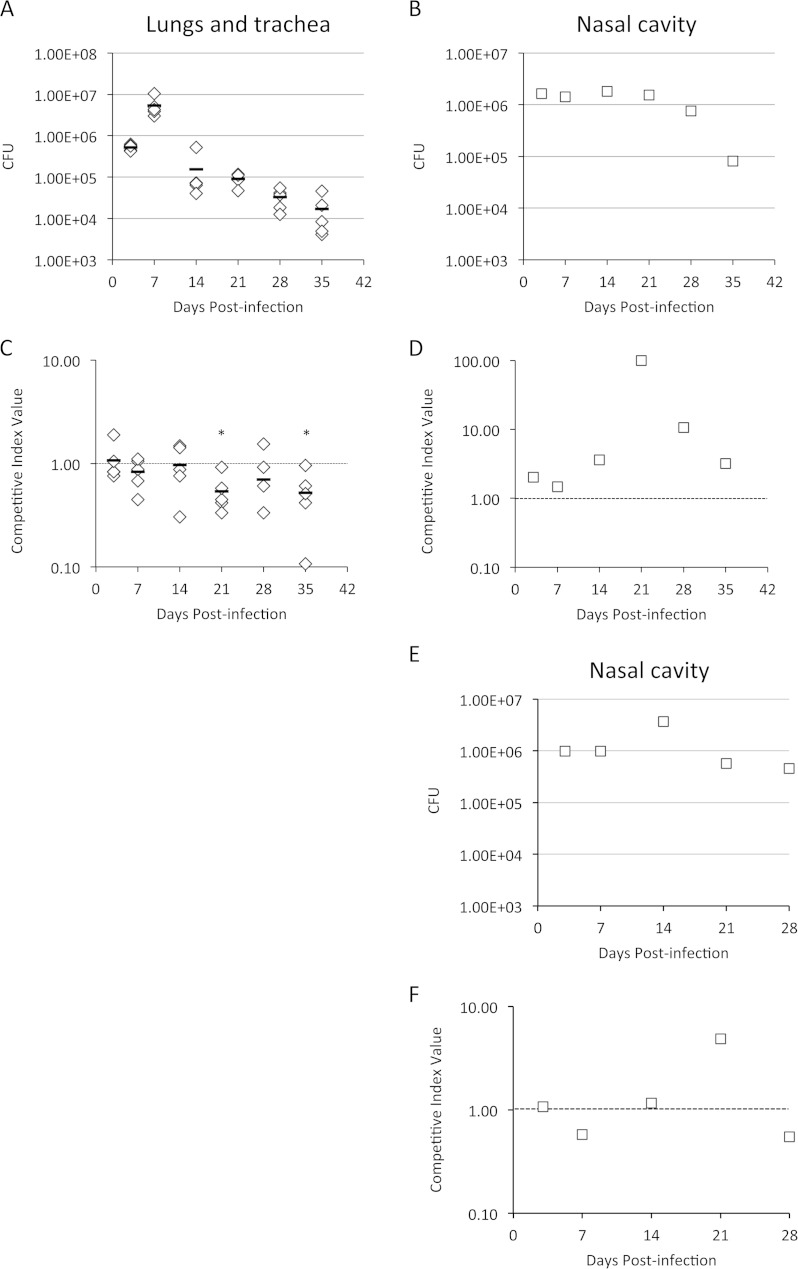
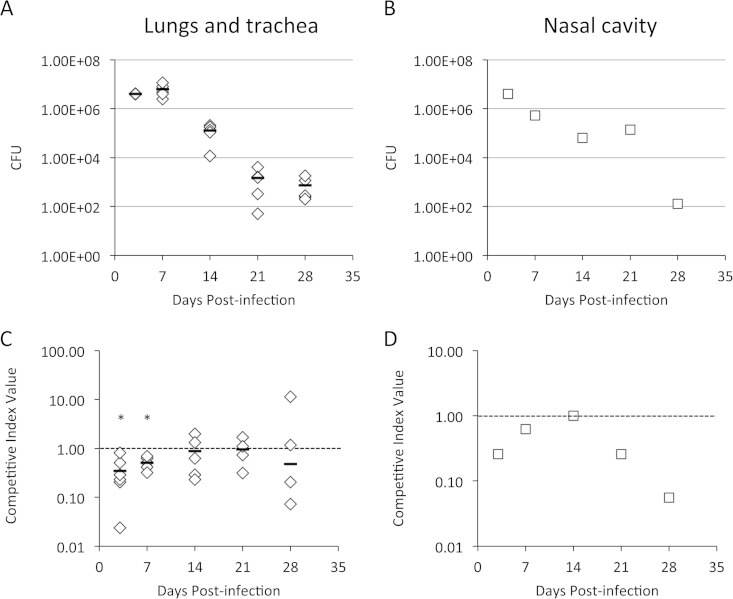
To determine if the BfrA, BfrD, and BfrE catecholamine receptors were important for growth in a host, mixed-infection competition experiments were performed. Mice were infected intranasally with a 1:1 mixture of B. bronchiseptica RB50 (wild-type strain) and RBB20 (isogenic bfrA bfrD bfrE mutant). At 3, 7, 14, 21, 28, and 35 days postinfection, infected mouse respiratory tract tissues were obtained, total and differential CFU counts were determined, and competitive index values were calculated (Fig. 8). Bacterial counts in the lungs and trachea (Fig. 8A) peaked at day 7 postinfection, gradually declining to 1.7 × 104 CFU per mouse by day 35. In these tissues, the B. bronchiseptica bfrA bfrD bfrE triple mutant exhibited little fitness defect until after day 14, in the later stages of infection (Fig. 8C). In the nasal cavity, bacterial counts hovered at ∼1.6 × 106 per animal until late infection, declining by approximately 1 log by day 35 (Fig. 8B). Interestingly, by midinfection (day 21), the mutant had an apparent fitness advantage in the nasal cavity (Fig. 8D).
FIG 8.
Relative in vivo fitness of a B. bronchiseptica bfrA bfrD bfrE triple receptor mutant during mouse respiratory infection. The graphs show the mean total CFU/mouse (n = 5) in the outputs recovered from lung and tracheal homogenates from individual mice (A) or from pooled nasal cavity homogenates (B and E) at various times postinfection. Nasal cavity data from two experiments are shown (experiment 1, panels B and D; and experiment 2, panels E and F). The lower limit of detection was 100 CFU per mouse. Competitive index (CI) values were calculated as the mutant/wild-type strain ratio in the outputs from lungs and tracheas of individual mice (C) or from pooled nasal cavities (D and F) recovered at each time point divided by the mutant/wild-type strain ratio in the input inoculum, as determined by differential colony hybridization. CI values are shown relative to the reference CI of 1.00 (dashed line). (C) For the lungs and trachea, the horizontal bars represent mean values, and each symbol represents the CI value determined for an individual mouse. P values of ≤0.05 (indicated with asterisks) were considered to be significant.
Since bfrD is activated by Bvg and expressed by B. pertussis in vitro and production of the BfrD protein has been reported for B. pertussis (22, 23, 47), mixed-infection competition experiments were performed to determine whether BfrD is important for growth of this Bordetella species in a host. Bacterial counts from the lungs and trachea peaked at day 7 and declined significantly by day 28 (Fig. 9A), which is characteristic of B. pertussis mouse infections. The B. pertussis bfrD mutant showed a modest but significant fitness defect in the lungs and trachea of mice early in infection (Fig. 9C). In the nasal cavity, CFU values similarly decreased over the infection period (Fig. 9B), and the mutant exhibited an apparently reduced fitness at early and late infection time points (Fig. 9D). Together, these results suggest a moderate yet significant role for the B. bronchiseptica and B. pertussis catecholamine receptors in this mouse model of infection.
FIG 9.
Relative in vivo fitness of a B. pertussis bfrD receptor mutant during mouse respiratory infection. The graphs show the mean total CFU/mouse (n = 5) in the outputs recovered from lung and tracheal homogenates from individual mice (A) or from pooled nasal cavity homogenates (B) at various times postinfection. The lower limit of detection was 100 CFU per mouse. CI values were calculated as described in the legend to Fig. 8, based on strain ratios determined by differential colony hybridization. CI values are shown relative to the reference CI of 1.00 (dashed line). (C) For the lungs and trachea, the horizontal bars represent mean values, and each symbol represents the CI value determined for an individual mouse.
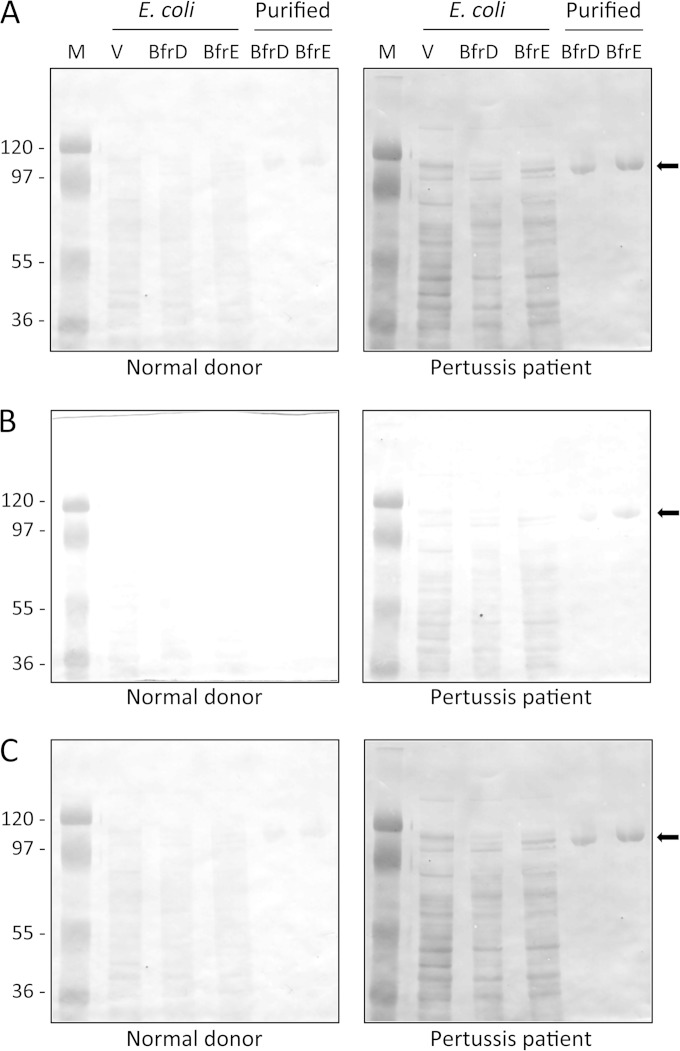
In a previous study from our laboratory, immunoblot screening of human sera from culture-positive patients diagnosed with pertussis demonstrated that, compared with control sera, the majority of infected patient sera demonstrated strong antibody reactivity to iron-repressible membrane proteins, including the B. pertussis outer membrane receptors for alcaligin, enterobactin, and heme (38). These human sera were used in similar analyses of B. pertussis BfrD and BfrE (Fig. 10). Evaluation of BfrD and BfrE produced in B. pertussis and B. bronchiseptica was complicated by the naturally low levels of receptor protein production. Therefore, B. pertussis bfrD and bfrE were expressed in E. coli, and the overproduced proteins were used as antigens for immunoblot analysis. Compared with sera from uninfected donors, sera from pertussis patients exhibited stronger reactivity (and consistently showed more background reactivity, similar to the results of our previous study [38]) with the recombinant BfrD and BfrE proteins. Multiple uninfected- and convalescent-phase-serum paired sample sets were analyzed in parallel, yielding similar results. The serum reactivity could also be due to antibodies that cross-react with other bacterial proteins, and there is likely cross-reactivity between the highly similar BfrD and BfrE proteins. However, overall, these results suggest that in the context of human infection, B. pertussis produces the BfrD and BfrE receptor proteins, which elicit an antibody response.
FIG 10.
Immunoblot reactivity of human sera with B. pertussis BfrD and BfrE receptor proteins. (A) Protein samples were analyzed in replicate immunoblots by using sera from an uninfected human donor (normal donor) and a B. pertussis culture-positive human donor (pertussis patient). Lanes: E. coli, total cell proteins prepared from E. coli producing recombinant BfrD and BfrE proteins; Purified, gel-purified authentic BfrD and BfrE proteins; M, molecular mass markers; V, cell proteins from E. coli carrying the plasmid vector control. (B) The antigens analyzed in panel A were probed using sera from donors different from those shown in panel A. (C) The antigens analyzed in panels A and B were probed using sera from donors different from those shown in panels A and B. Positions of BfrD and BfrE are indicated by arrows.
DISCUSSION
Studies from this laboratory demonstrated B. bronchiseptica utilization of transferrin- and lactoferrin-bound iron via the catecholamines epinephrine, norepinephrine, dopamine, and l-3,4-dihydroxyphenylalanine (l-DOPA), as well as 2,5-dihydroxybenzoylserine (20). BfrA, BfrD, and BfrE were identified as outer membrane receptors involved in this iron acquisition mechanism, which operates in the absence of siderophores. Catecholamines act as chelators that effectively remove the iron from transferrin or lactoferrin (53). Catecholamine-iron complexes are predicted to diffuse to the Bordetella cell surface and interact with BfrA, BfrD, and/or BfrE for uptake to the periplasm. The machinery that transports the iron across the cytoplasmic membrane to the cytoplasm is not known, and there are no obvious candidate transporter genes colocalized on the chromosome near bfrA, bfrD, or bfrE.
Taxonomically related Bordetella petrii and Achromobacter species that live in the external environment have a single gene encoding a protein that is significantly more similar to BfrE (80% identity) than to BfrD (56 to 57% identity). This suggests that bfrE was duplicated in the classical pathogenic Bordetella species, giving rise to the adjacent bfrD gene, which evolved to be coregulated with virulence factor genes via BvgAS. B. pertussis has intact bfrD and bfrE genes, but demonstration of catecholamine-mediated growth on transferrin or lactoferrin iron by B. pertussis has not been successful with our in vitro culture system. However, the present study showed that the B. pertussis BfrD and BfrE proteins are functional for catecholamine utilization when the genes are expressed in B. bronchiseptica, illustrating differences between the human-adapted species and B. bronchiseptica. In our studies of Bordetella iron acquisition, we observed no significant differences in iron requirements or sensitivity to iron chelators between B. bronchiseptica and B. pertussis that may account for the differences in catecholamine receptor function. It is possible that our in vitro culture system using catecholamines and ferric transferrin does not recapitulate conditions allowing BfrD/BfrE function in B. pertussis. It is also possible that B. pertussis never acquired, or has lost, the other factors for catecholamine utilization, such as a cytoplasmic membrane transporter. However, previous studies showed that the B. bronchiseptica bfrA gene could promote norepinephrine-mediated utilization of transferrin iron by B. pertussis, which lacks a bfrA ortholog (20), suggesting that a catecholamine transporter system that can function with BfrA is active in B. pertussis. Database searches indicate that all sequenced B. pertussis strains have intact bfrD and bfrE genes, and BfrD was proposed as a potential vaccine antigen (23). Perhaps, in contrast to the case in B. bronchiseptica, B. pertussis BfrD/BfrE function continued to evolve in humans such that noncatecholamine host substrates were bound and transported. Our infection experiments indicate that BfrD does have an apparent function in B. pertussis, since a bfrD mutant is attenuated in mice, albeit modestly. Our studies also indicate that sera from humans convalescing from pertussis recognize the BfrD and BfrE proteins, suggesting the in vivo expression of bfrD and bfrE in B. pertussis in the natural human host. Previously, we demonstrated convalescent-phase human serum reactivity to a variety of membrane proteins from iron-starved (versus iron-replete) Bordetella cells, including the TonB-dependent receptors for alcaligin, heme, and enterobactin (38).
Previous studies from this laboratory showed that B. pertussis mutants lacking the receptors for alcaligin (FauA), heme (BhuR), and enterobactin (BfeA) were attenuated for growth in the mouse respiratory tract (35, 36, 38). In the present study, our results demonstrated that a B. pertussis bfrD mutant was less fit than the wild-type parent strain in the lungs and trachea, especially during the first week of infection. Comparison of competitive index values from these experiments indicates that the bfrD mutant had a smaller fitness defect than those of the fauA and bhuR mutants but a larger fitness defect than that of the bfeA mutant. In the lungs and trachea, a B. bronchiseptica bfrA bfrD bfrE triple receptor mutant exhibited decreased fitness in later stages of infection. Unexpectedly, the B. bronchiseptica competitive index in the nasal cavity suggested that the triple mutant had a fitness advantage over the wild-type strain, especially at day 21. Although significance cannot be determined because the nasal samples were pooled, similar nasal fitness results were obtained in another experiment. The B. pertussis bfrD mutant showed a fitness defect in the nasal cavity at both early and later infection time points, but unlike B. bronchiseptica, it did not exhibit any fitness advantage over the coinfecting wild-type strain. During our in vitro analyses of B. bronchiseptica bfrA, bfrD, and/or bfrE single- or multiple-receptor mutants, we did not observe overproduction of the alcaligin siderophore or any other effects on siderophore-mediated iron uptake that may explain the apparent nasal fitness advantage. A recent study examined mouse nasal colonization efficacies of B. pertussis and B. bronchiseptica and showed significant species differences that involved the respiratory microbiota (54). B. bronchiseptica nasal colonization was obtained using infecting doses as low as 5 CFU, whereas much higher doses of B. pertussis (10,000 CFU) or antibiotic pretreatment was required to overcome inhibition by the microflora. Our observed species differences in the mouse nasal cavity may reflect these known effects of the resident flora. The Bordetella species growth differences in the mouse respiratory tract as a whole also may reflect differences in catecholamine receptor functions. In addition, compared with B. pertussis, B. bronchiseptica generally causes chronic infection in its natural nonhuman mammalian hosts, such as mice (2, 4, 55), and one or more catecholamine receptors may play a role in persistence.
In previous transcriptome analyses of iron-starved versus iron-replete B. pertussis and B. bronchiseptica, differences in bfrA, bfrD, and bfrE expression levels did not meet the cutoff value to allow their assignment as either iron repressed or iron induced (44). In the present study, we found that bfrA and bfrE expression levels were low under all growth conditions. A previous study also reported very low expression of bfrE-lacZ in B. pertussis (21). BfrD was originally described as an outer membrane protein produced by Bvg+ virulent-phase B. pertussis (22), and a subsequent mutagenesis study using lacZ gene fusions confirmed the Bvg regulation of B. pertussis bfrD (21). In those studies, the bacteria were cultured in iron-replete medium, since the functions of BfrD and BfrE were unknown at the time. In the present analysis, bfrE and bfrD exhibited higher transcription levels when bacteria were grown in iron-replete medium than in iron-depleted SS medium. Typically, iron acquisition genes are repressed by Fur and iron under iron-replete growth conditions, and thus show elevated expression during iron starvation. For bfrE in both Bordetella species, expression under both iron conditions was very low. In contrast, bfrD transcription was consistently and significantly higher in iron-replete medium than the expression in iron-starved B. bronchiseptica and B. pertussis. Iron starvation is an important cue for pathogenic bacteria, informing them that they are in the host environment, where free iron is scarce (56, 57). It is possible that having bfrD under BvgAS virulence control serves the same signaling purpose in Bordetella cells, supplanting the role of the iron starvation cue. However, since iron-replete in vitro growth conditions increased bfrD transcription, it may be that other signals feed into the regulatory pathway in the host environment.
Alternatively, other mechanisms, such as differential mRNA stability or small regulatory RNA species, may control bfrD transcription in relationship to the organism's iron status. Although regulatory RNAs have been found in B. pertussis, those controlling TonB-dependent receptor gene expression have not been reported (58).
During modulation, Bordetella cells are in the Bvg− phenotypic phase; Bvg-activated virulence genes are minimally transcribed, and genes such as those involved in motility and chemotaxis are maximally expressed. The Bvg intermediate-phase genes are maximally expressed at intermediate modulator concentrations (2, 4). When Bordetella cells are switched from modulating conditions to nonmodulating conditions, the transit from the Bvg− to Bvg+ phase is characterized by restoration of Bvg-activated gene expression, paralleling the increasing intracellular phosphorylated BvgA (BvgA∼P) concentration. Expression of the class 2 genes (e.g., those encoding fimbriae, filamentous hemagglutinin, and BvgAS) resumes early after removal of modulators, and transcription of the class 1 genes (encoding pertussis toxin, adenylate cyclase, and the type III secretion system) resumes late in this process (59). A previous study noted that the cyaA adenylate cyclase gene, a class 1 late gene in B. pertussis, was transcribed significantly earlier in B. bronchiseptica (60). There is significant variability in the BvgS amino acid sequences of the classical Bordetella species (52, 61, 62). Using the B. bronchiseptica RB52 strain carrying the B. pertussis bvgAS genes, previous studies indicated that this difference in cyaA expression was not due to species differences in the cyaA promoter or bvgAS sequences (60). The authors postulated that the cyaA transcription profiles were likely due to other differences between the two Bordetella species. Our results demonstrated variations in bfrD expression between B. pertussis and B. bronchiseptica. Notably, B. bronchiseptica bfrD expression was resistant to modulation by nicotinic acid and sulfate, whereas in B. pertussis, the same gene was readily modulated, as was native B. pertussis bfrD. In strain RB52 carrying bvgASBp, bfrD modulation was achieved only at the highest modulator concentrations, compared with no modulation in the wild-type parent strain RB50. These results indicate that the Bordetella species differences observed in bfrD transcription may be due to variations in BvgAS. It is clear that expression of bfrD in both Bordetella species requires BvgAS. However, in B. bronchiseptica, the relative resistance of bfrD to modulation indicates that very low concentrations of BvgA∼P are sufficient for transcriptional activation and high-level expression of the gene.
As obligate mammalian pathogens, the classical Bordetella species evolved to utilize host nutritional resources. In the host respiratory tract, it is unknown whether catecholamines are the authentic substrates for the BfrA, BfrD, and BfrE receptors. Serum is known to exude onto the respiratory mucosa (63), and metabolomic studies of serum indicate the presence of not only catecholamines but also catechols and other aromatic compounds (64), which may also be substrates for the receptors. Given the differences in B. pertussis and B. bronchiseptica host ranges and the acute versus chronic nature of the infections they cause, their catecholamine receptors may have evolved to carry out distinct functions.
ACKNOWLEDGMENTS
This work was supported by U.S. Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
We thank Andrew Norgan for construction of plasmid pMP2.
REFERENCES
- 1.Bordet J, Gengou O. 1906. Le microbe de la coqueluche. Ann Inst Pasteur 20:731. [Google Scholar]
- 2.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherry JD. 2012. Epidemic pertussis in 2012—the resurgence of a vaccine-preventable disease. N Engl J Med 367:785–787. doi: 10.1056/NEJMp1209051. [DOI] [PubMed] [Google Scholar]
- 4.Melvin JA, Scheller EV, Miller JF, Cotter PA. 2014. Bordetella pertussis pathogenesis: current and future challenges. Nat Rev Microbiol 12:274–288. doi: 10.1038/nrmicro3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss A, Hewlett E, Myers G, Falkow S. 1983. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect Immun 42:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aricó B, Miller JF, Roy C, Stibitz S, Monack D, Falkow S, Gross R, Rappuoli R. 1989. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc Natl Acad Sci U S A 86:6671–6675. doi: 10.1073/pnas.86.17.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg ED. 1978. Iron and infection. Microbiol Rev 42:45–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullen J. 1981. The significance of iron in infection. Rev Infect Dis 3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 9.Brickman TJ, Armstrong SK. 2009. Temporal signaling and differential expression of Bordetella iron transport systems: the role of ferrimones and positive regulators. Biometals 22:33–41. doi: 10.1007/s10534-008-9189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postle K, Larsen RA. 2007. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- 11.Parkhill J, Sebaihia M, Preston A, Murphy L, Thomson N, Harris D, Holden M, Churcher C, Bentley S, Mungall K, Cerdeno-Tarraga A, Temple L, James K, Harris B, Quail M, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell B, Maskell D. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 12.Anderson MT, Armstrong SK. 2008. Norepinephrine mediates acquisition of transferrin-iron in Bordetella bronchiseptica. J Bacteriol 190:3940–3947. doi: 10.1128/JB.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore CH, Foster LA, Gerbig DG, Dyer DW, Gibson BW. 1995. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J Bacteriol 177:1116–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brickman TJ, Hansel JG, Miller MJ, Armstrong SK. 1996. Purification, spectroscopic analysis and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. Biometals 9:191–203. [DOI] [PubMed] [Google Scholar]
- 15.Beall B, Sanden GN. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- 16.Beall B, Hoenes T. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143:135–145. doi: 10.1099/00221287-143-1-135. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson ML, Beall B. 1999. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology 145:2453–2461. doi: 10.1099/00221287-145-9-2453. [DOI] [PubMed] [Google Scholar]
- 18.Pradel E, Guiso N, Menozzi FD, Locht C. 2000. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect Immun 68:1919–1927. doi: 10.1128/IAI.68.4.1919-1927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanderpool CK, Armstrong SK. 2001. The Bordetella bhu locus is required for heme iron utilization. J Bacteriol 183:4278–4287. doi: 10.1128/JB.183.14.4278-4287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong SK, Brickman TJ, Suhadolc RJ. 2012. Involvement of multiple distinct Bordetella receptor proteins in the utilization of iron liberated from transferrin by host catecholamine stress hormones. Mol Microbiol 84:446–462. doi: 10.1111/j.1365-2958.2012.08032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoine R, Alonso S, Raze D, Coutte L, Lesjean S, Willery E, Locht C, Jacob-Dubuisson F. 2000. New virulence-activated and virulence-repressed genes identified by systematic gene inactivation and generation of transcriptional fusions in Bordetella pertussis. J Bacteriol 182:5902–5905. doi: 10.1128/JB.182.20.5902-5905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passerini de Rossi BN, Friedman LE, Gonzalez Flecha FL, Castello PR, Franco MA, Rossi JP. 1999. Identification of Bordetella pertussis virulence-associated outer membrane proteins. FEMS Microbiol Lett 172:9–13. doi: 10.1111/j.1574-6968.1999.tb13442.x. [DOI] [PubMed] [Google Scholar]
- 23.de Gouw D, de Jonge MI, Hermans P. 2014. Proteomics-identified Bvg-activated autotransporters protect against Bordetella pertussis in a mouse model. PLoS One 9:e105011. doi: 10.1371/journal.pone.0105011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stainer D, Scholte M. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol 63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 25.Schneider D, Parker C. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect Immun 38:548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 27.Figurski D, Helinski D. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 29.Spaink HP, Okker R, Wijffelman CA, Pees E, Lugtenberg B. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid Prl1ji. Plant Mol Biol 9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 30.Karimova G, Ullmann A. 1997. Characterization of DNA binding sites for the BvgA protein of Bordetella pertussis. J Bacteriol 179:3790–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brickman TJ, Armstrong SK. 1996. Colicins B and Ia as novel counterselective agents in interspecies conjugal DNA transfers from colicin-sensitive Escherichia coli donors to other gram-negative recipient species. Gene 178:39–42. doi: 10.1016/0378-1119(96)00331-9. [DOI] [PubMed] [Google Scholar]
- 32.Miller JH. 1972. Assay of β-galactosidase, p 352–355. In Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 33.Stibitz S. 1994. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol 235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 34.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 35.Brickman TJ, Vanderpool CK, Armstrong SK. 2006. Heme transport contributes to in vivo fitness of Bordetella pertussis during primary infection in mice. Infect Immun 74:1741–1744. doi: 10.1128/IAI.74.3.1741-1744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brickman TJ, Armstrong SK. 2007. Impact of alcaligin siderophore utilization on in vivo growth of Bordetella pertussis. Infect Immun 75:5305–5312. doi: 10.1128/IAI.00849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mobberley-Schuman PS, Connelly B, Weiss AA. 2003. Phagocytosis of Bordetella pertussis incubated with convalescent serum. J Infect Dis 187:1646–1653. doi: 10.1086/374741. [DOI] [PubMed] [Google Scholar]
- 38.Brickman TJ, Hanawa T, Anderson MT, Suhadolc RJ, Armstrong SK. 2008. Differential expression of Bordetella pertussis iron transport system genes during infection. Mol Microbiol 70:3–14. doi: 10.1111/j.1365-2958.2008.06333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller A, Nesvizhskii A, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 40.Nesvizhskii A, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 41.Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 42.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 43.Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brickman TJ, Cummings CA, Liew S-Y, Relman DA, Armstrong SK. 2011. Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J Bacteriol 193:4798–4812. doi: 10.1128/JB.05136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calderwood SB, Mekalanos JJ. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol 169:4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Lorenzo V, Wee S, Herrero M, Neilands JB. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol 169:2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Passerini de Rossi BN, Friedman LE, Belzoni CB, Savino S, Aricò B, Rappuoli R, Masignani V, Franco MA. 2003. Vir90, a virulence-activated gene coding for a Bordetella pertussis iron-regulated outer membrane protein. Res Microbiol 154:443–450. doi: 10.1016/S0923-2508(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 48.Stojiljkovic I, Baumler AJ, Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol 236:531–545. [DOI] [PubMed] [Google Scholar]
- 49.Brickman TJ, Armstrong SK. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J Bacteriol 181:5958–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanderpool CK, Armstrong SK. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J Bacteriol 185:909–917. doi: 10.1128/JB.185.3.909-917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson MT, Armstrong SK. 2004. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J Bacteriol 186:7302–7311. doi: 10.1128/JB.186.21.7302-7311.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Tejada GM, Miller JF, Cotter PA. 1996. Comparative analysis of the virulence control systems of Bordetella pertussis and Bordetella bronchiseptica. Mol Microbiol 22:895–908. doi: 10.1046/j.1365-2958.1996.01538.x. [DOI] [PubMed] [Google Scholar]
- 53.Sandrini SM, Shergill R, Woodward J, Muralikuttan R, Haigh RD, Lyte M, Freestone PP. 2010. Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin. J Bacteriol 192:587–594. doi: 10.1128/JB.01028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weyrich LS, Feaga HA, Park J, Muse SJ, Safi CY, Rolin OY, Young SE, Harvill ET. 2014. Resident microbiota affect Bordetella pertussis infectious dose and host specificity. J Infect Dis 209:913–921. doi: 10.1093/infdis/jit597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodnow RA. 1980. Biology of Bordetella bronchiseptica. Microbiol Rev 44:722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahan M, Heithoff D, Sinsheimer R, Low D. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu Rev Genet 34:139–164. doi: 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]
- 57.Conway T, Schoolnik GK. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol Microbiol 47:879–889. doi: 10.1046/j.1365-2958.2003.03338.x. [DOI] [PubMed] [Google Scholar]
- 58.Hot D, Slupek S, Wulbrecht B, D'Hondt A, Hubans C, Antoine R, Locht C, Lemoine Y. 2011. Detection of small RNAs in Bordetella pertussis and identification of a novel repeated genetic element. BMC Genomics 12:207. doi: 10.1186/1471-2164-12-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cotter P, Jones AM. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol 11:367–373. doi: 10.1016/S0966-842X(03)00156-2. [DOI] [PubMed] [Google Scholar]
- 60.Byrd MS, Mason E, Henderson MW, Scheller EV, Cotter PA. 2013. An improved recombination-based in vivo expression technology-like reporter system reveals differential cyaA gene activation in Bordetella species. Infect Immun 81:1295–1305. doi: 10.1128/IAI.01445-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aricó B, Scarlato V, Monack DM, Falkow S, Rappuoli R. 1991. Structural and genetic analysis of the bvg locus in Bordetella species. Mol Microbiol 5:2481–2491. doi: 10.1111/j.1365-2958.1991.tb02093.x. [DOI] [PubMed] [Google Scholar]
- 62.Herrou J, Debrie A-S, Willery E, Renaud-Mongénie G, Locht C, Mooi F, Jacob-Dubuisson F, Antoine R. 2009. Molecular evolution of the two-component system BvgAS involved in virulence regulation in Bordetella. PLoS One 4:e6996. doi: 10.1371/journal.pone.0006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Persson CG, Erjefalt JS, Greiff L, Andersson M, Erjefält I, Godfrey RW, Korsgren M, Linden M, Sundler F, Svensson C. 1998. Plasma-derived proteins in airway defence, disease and repair of epithelial injury. Eur Respir J 11:958–970. doi: 10.1183/09031936.98.11040958. [DOI] [PubMed] [Google Scholar]
- 64.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. 2011. The human serum metabolome. PLoS One 6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kasuga T, Nakase Y, Ukishima K, Takatsu K. 1954. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch Exp Med 27:57–62. [PubMed] [Google Scholar]
- 66.Field L, Parker C. 1979. Differences observed between fresh isolates of Bordetella pertussis and their laboratory-passaged derivatives, p 124–132. In Proceedings of the International Symposium on Pertussis. U.S. Department of Health, Education, and Welfare, Washington, DC. [Google Scholar]
- 67.Brickman TJ, Armstrong SK. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp: putrescine is a precursor of alcaligin. J Bacteriol 178:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cotter P, Miller J. 1994. BvgAS-mediated signal transduction—analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun 62:3381–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farrington DO, Switzer WP. 1979. Parenteral vaccination of young swine against Bordetella bronchiseptica. Am J Vet Res 40:1347–1351. [PubMed] [Google Scholar]
- 70.Armstrong SK, Clements MO. 1993. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J Bacteriol 175:1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brickman TJ, Armstrong SK. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J Bacteriol 187:3650–3661. doi: 10.1128/JB.187.11.3650-3661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]