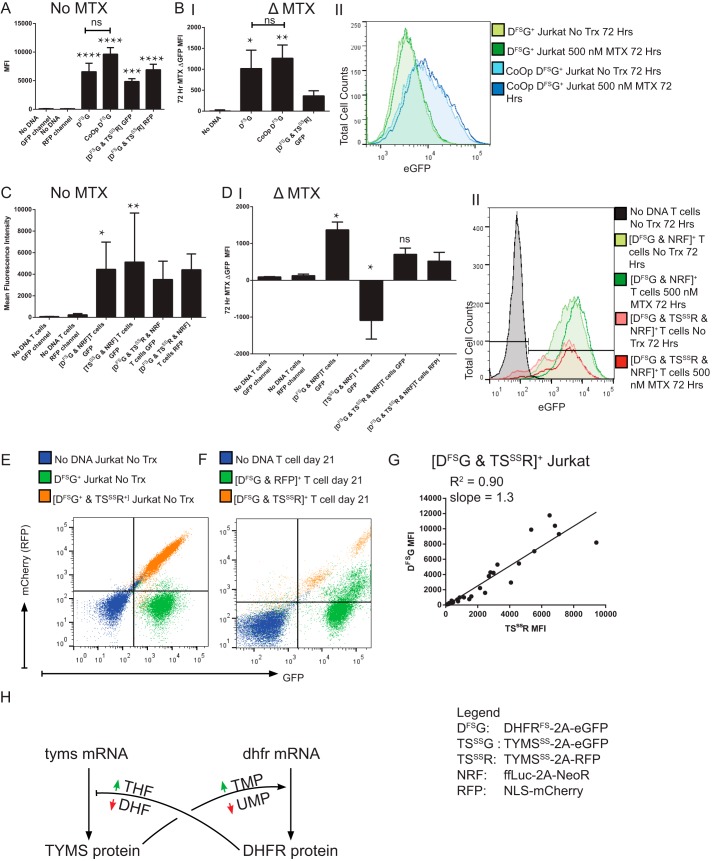
FIGURE 1.
DHFRFS 3′ cis-transgene expression increases in the presence of MTX independent of mRNA sequence, and the increase is suppressed by restoration of thymidine synthesis. A, Jurkat cells were genetically modified to express FLAG-DHFRFS-2A-eGFP pSBSO (DFSG) with resistance to MTX (n = 4), CoOp DFSG, with known mRNA binding elements DFSG removed (n = 5), and [DFSG & FLAG-TYMSSS-2A-RFP pSBSO (TSSSR)], with enhanced resistance to MTX beyond DFSG alone through the addition of MTX-resistant TYMSSS (n = 7). Genetically modified Jurkat cells were selected for 2 weeks in 1 μm MTX before culturing without MTX for 3–5 weeks. In A, the stable fluorescent protein expression, in the absence of MTX, is depicted by MFI. B, panel I, Jurkat cells were treated for 72 h with 0.5 μm MTX or no treatment. The MFI difference (Δ = eGFP MFI MTX-treated − eGFP MFI-untreated) is depicted. B, panel II, a histogram representative of the data used in panel I demonstrates the MTX-induced change in eGFP MFI for DHFRFS and CoOp DHFRFS in Jurkat cells. No Trx, no treatment. C, in primary T cells, transgenes DHFRFS, TYMSSS, or the combination were selected with ffLuc-NeoR pSBSO (NRF) for 2 weeks in the presence of cytotoxic drug and then propagated without selection for 3 weeks (see “Experimental Procedures”). On day 35, T cells were stimulated with anti-CD3, anti-CD28 antibodies, and 50 IU/ml IL-2 in the presence or absence of MTX for 72 h. The fluorescent protein MFI of untreated cells is shown. D, panel I, depicts the Δ MFI after 72 h of treatment with 0.5 μm MTX in comparison to no treatment. In D, panel II, a representative histogram demonstrates the observed shift in eGFP fluorescence for DHFRFS+ T cells in the presence or absence of MTX (n = 5). E, a trans regulatory pattern of DHFR- and TYMS-linked proteins was observed. A representative flow plot from the 1 μm MTX-selected Jurkat cells left untreated, as in A, demonstrates that in untreated Jurkat cells of either unselected mock-electroporated (No DNA) or DFSG+, there is a globular appearance in the RFP channel. However, co-expression of DHFRFS with TYMSSS in [DFSG & TSSSR]+ Jurkat cells leads to a linear clustering. F, primary T cells were electroporated with DHFRFS and co-transformed with either RFP control or FLAG-TYMSSS-2A-RFP pSBSO (TSSSR) before propagation, as in C. A representative flow plot of primary human T cells from the same donor where [DFSG & RFP], [DFSG & TSSSR], and untransformed T cells is shown on day 21, after 7 days without selection in MTX. A linear clustering of DHFRFS is again noted when co-expressed with TYMSSS that is not noted with RFP alone. G, further studies to identify a trans pattern of linked expression between DHFRFS and TYMSSS were identified in the selection of [DFSG & TSSSR] electroporated Jurkat cells in antifolates MTX (0, 0.01, 0.1, 0.5, 1, 5 μm), pemetrexed (0, 10, 50, 100 μm), and raltitrexed (0, 1, 5, 10 μm). The MFI of DFSG and TSSSR for each expression pattern was plotted after days 2–14 in selection. The values were plotted, and a linear fitting was performed with the R2 from the Pearson's correlation and the slope of the linear regression provided on the graph. These data are assembled from four technical replicates. H, a model of post-transcriptional regulation of DHFR and TYMS mRNA into the respective proteins is depicted with colored arrows indicating increase (green) or decrease (red) in specific small molecules. Black pointed arrows indicate promotion, while black bars indicate inhibition of protein translation. All experiments other than G were independently repeated twice. Kruskal-Wallis test was used to determine significant differences with multivariate analyses. Unmarked statistically significant results refer to comparison between experimental cells and the “No DNA” untreated control cells of equivalent measure. ns = not significant; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. TMP, thymidine monophosphate; UMP, uridine monophosphate; DHF, dihydrofolate.