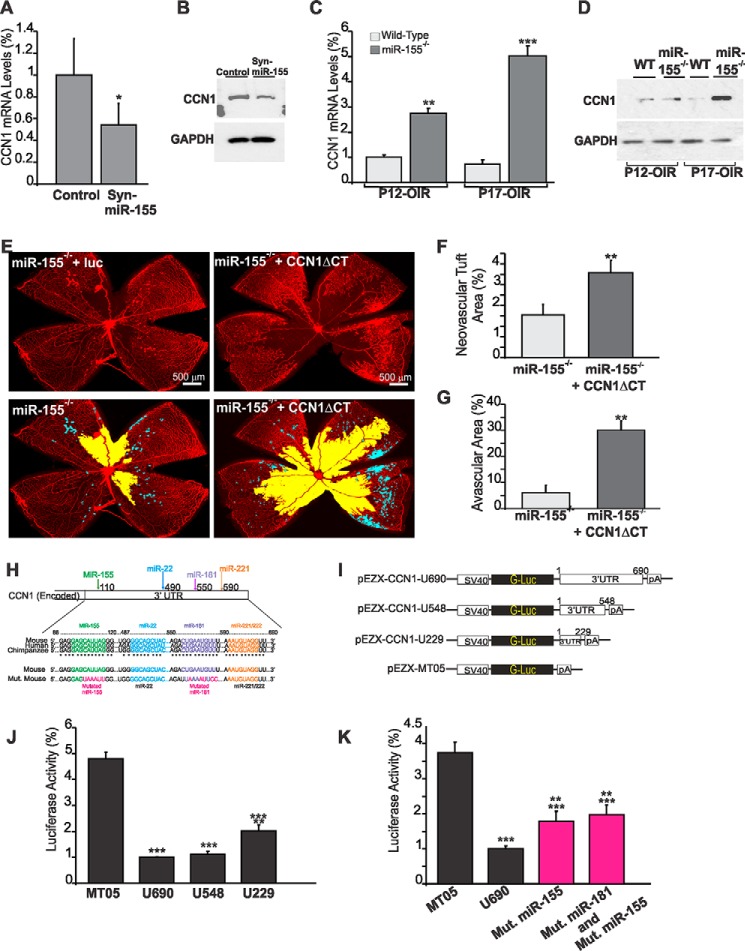
FIGURE 4.
MicroRNA-155 directly targets CCN1 gene expression during vessel and neovessel growth. A and B, effects of Syn-miR-155 injection on CCN1 gene expression during retinal vessel development. CCN1 gene expression was determined at the mRNA (A) and protein level (B) in retinal tissue lysates from wild-type mice at P6 after treatment at P3 with either scrambled control miRNA (Cntl) or Syn-miR-155. *, p < 0.05 (n = 3). C and D, expression of the CCN1 gene at the mRNA (C) and protein (D) levels in wild-type and miR-155-deficient mice following OIR. Wild-type and miR-155−/− mice were placed in 75% oxygen at P7 and returned to room air at P12. Retinas were harvested at P12 or P17, and CCN1 expression was analyzed by either qPCR or Western immunoblotting as described under “Experimental Procedures.” CCN1 mRNA levels were normalized to those of acidic ribosomal phosphoprotein. **, p < 0.01 versus wild type. ***, p < 0.001 versus wild type (n = 5). GAPDH was used as a loading control of protein lysate analysis. E–G, effects of CCN1 suppression on the vascularization and neovascularization of the retina of miR-155−/− mice following OIR. Whole mount retinas shown were from eyes injected at P4 with either lnv-luc or lnv-CCN1ΔCT encoding a dominant negative form of CCN1. Mice were subjected to OIR at P7. Areas of vaso-obliteration and preretinal neovascular tufts at P17 as determined by computer-assisted image analyses are shown in yellow and blue, respectively (E). Compiled data showing the percentage of neovascular and avascular areas in eyes injected with lnv-luc and lnv-CCN1ΔCT are shown in F and G, respectively. **, p < 0.01 versus miR-155−/− (n = 5). H, schematic layout of the CCN1 3′-UTR, with the relative location of miR-155, miR-22, miR-181, and miR-221 binding sites. Depiction is not to scale. 3′-UTR sequences of CCN1 of mouse, human, and chimpanzee and predicted interaction with conserved miRNA seeds are shown. The sequence of the CCN1 3′-UTR seed mutant used for reporter assays and predicted disruption of the miR-155 and miR-181 interactions are also shown. I–K, transient transfection of endothelial cells was performed with an empty plasmid (MT05) or pEZX-CCN1 3′-UTR plasmids with and without mutated miRNA sequences depicted in H. Luciferase activity assays were performed on the supernatant media as described under “Experimental Procedures.” Gaussia luciferase values have been normalized to those of Cypridina luciferase, and the percentage of luciferase activity is represented. **, p < 0.05 versus U690. ***, p < 0.001 versus MT05 (n = 4). Error bars, S.E.