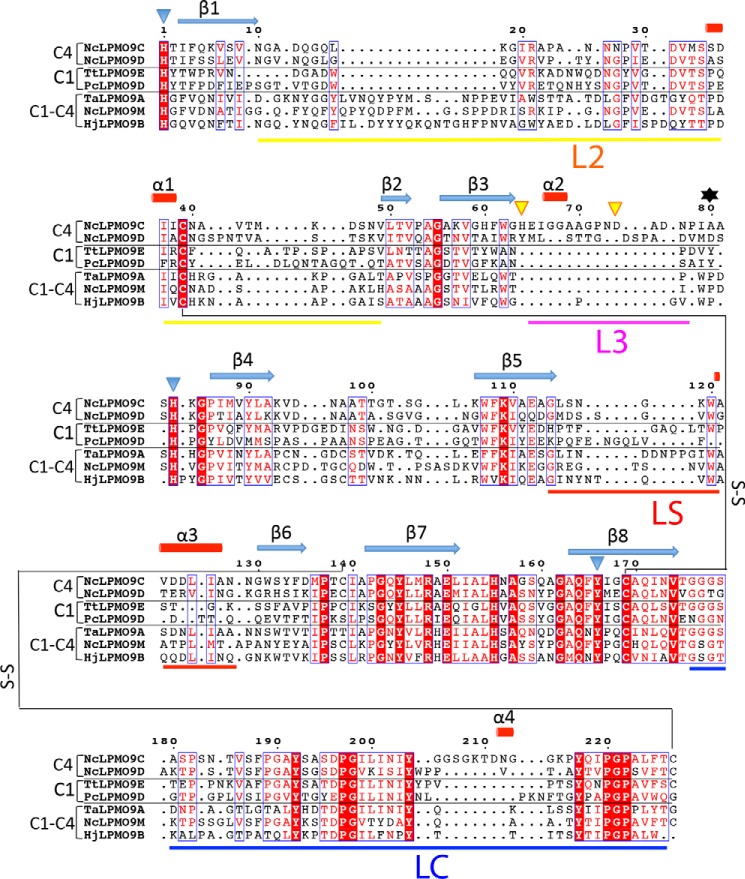
FIGURE 2.
Structure-based sequence alignment of LPMO9s with known structures. The proteins included are as follows: NcLPMO9C-N (PDB code 4D7U), NcLPMO9D (PDB code 4EIR), TtLPMO9E (PDB code 3EII), PcLPMO9D (PDB code 4B5Q), TaLPMO9A (PDB code 3ZUD), NcLPMO9M (PDB code 4EIS), and HjLPMO9B (PDB code 2VTC). Fully conserved residues are shown in white on a red background. Blue frames indicate that more than 70% of the residues in the corresponding columns exhibit similar physico-chemical properties (indicated as red residues on a white background). Blue triangles indicate residues coordinating the active site metal, and yellow triangles indicate residues involved in binding of two additional zinc ions. The secondary structure assignment (β-strands indicated as blue arrows and α-helices as red cylinders) refers to NcLPMO9C-N and was determined with the program DSSP (56). The oxidative regio-specificity of the LPMOs, indicated on the left, was assigned based on experimental evidence (4, 5, 13, 57) or, for HjLPMO9B and TtLPMO9E, by inference from the sequence-based categorization (43, 45). The residue numbered 80, which affects the accessibility of the solvent-facing axial copper coordination site, as shown in Fig. 4, c and d, and discussed in detail in the text, is indicated by a black asterisk. The loop regions that contribute to shaping the substrate-binding surface, named L2, LC, L3, and LS (see text), are marked by horizontal bars below the sequence, with color coding as in Fig. 2. The figure was prepared with ESPript.