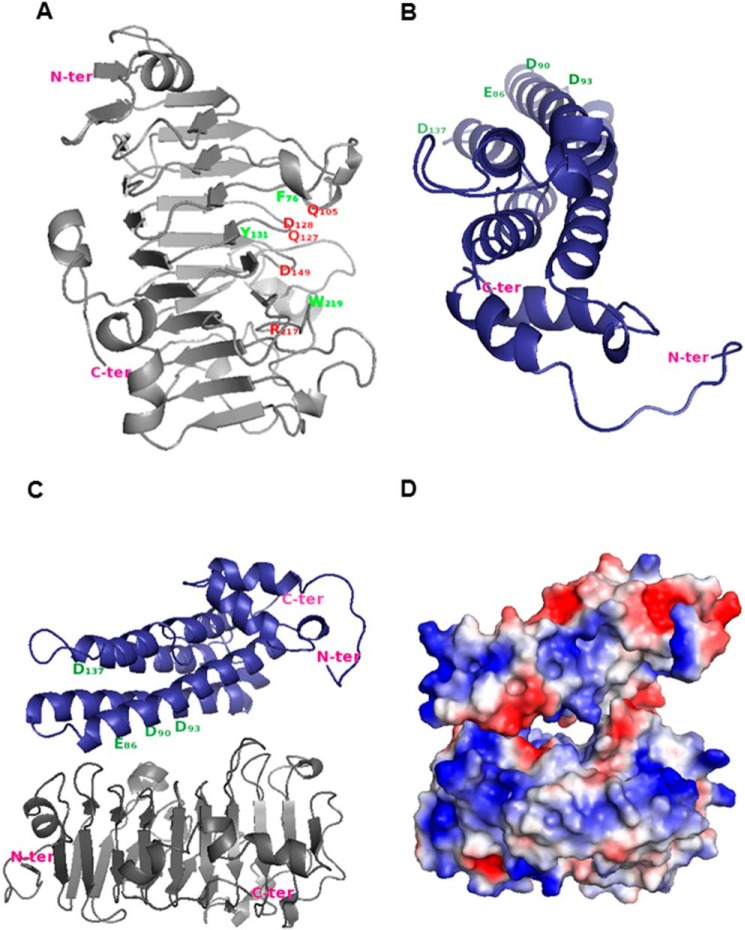
FIGURE 7.
Proposed model for the inactivation of PME3 by PMEI7. A, PME3 model. Putative amino acid residues involved in the catalytic site are shown in red, and putative important residues involved in both the pectin-binding site and the PME-PMEI interaction are shown in green. B, PMEI7 model. Putative amino acid residues interacting with the PME pectin-binding site are shown in green. Arabidopsis PME3 and PMEI7 models were built using carrot PME (PDB code 1GQ8, chain A) and kiwi PMEI (PDB code 1XG2, chain B) coordinates, respectively. The tertiary structure was modeled according to Ref. 31. C, interaction between PME3 (gray) and PMEI7 (blue) was evaluated as described (55, 56) using LZerD based on shape complementarity. Model number 8 from the 15 best models was selected based on the literature data and the number of contacting residues. D, electrostatic potential of PMEI7 on PME3.