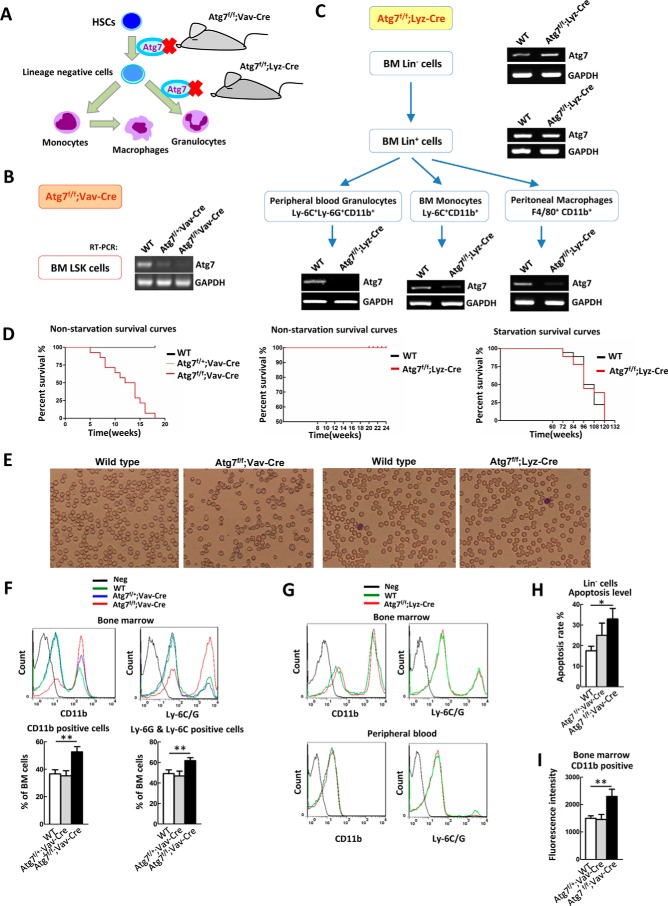
FIGURE 1.
Differential deletion of Atg7 in stem cells and terminally differentiated cells of hematopoietic system reveals different phenotypes. A, scheme for deferential deletion of atg7 in mouse hematopoietic hierarchy. B, identification of Atg7f/f;Vav-Cre mice. Atg7 gene is deleted in bone marrow LSK hematopoietic stem and progenitor cells. C, identification of Atg7f/f;Lyz-Cre mice. Atg7 gene is deleted in the myeloid cells including granulocytes, monocytes, and macrophages, but their upstream cells (bone marrow lineage negative and positive cells) maintain atg7 transcript. The designated types of cells from the mouse models were directly sorted from bone marrow cells by FACS, and the presence of Atg7 transcript was determined by RT-PCR. D, Atg7 deletion in hematopoietic stem cells, not myeloid cells, caused mouse death, but atg7 deletion in myeloid cells did not affect mouse death under nutrient-rich or starvation conditions. For starvation, mice were provided with water but without feed. E, blood routine examination of Atg7f/f;Vav-Cre mice and Atg7f/f;Lyz-Cre mice. F, flow cytometric analysis of the percentage of bone marrow myeloid cells (CD11b+) and granulocyte (Ly-6G+ and Ly-6C+) in Atg7f/f;Vav-Cre and wild-type mice. G, flow cytometric analysis of bone marrow and peripheral blood myeloid cells (CD11b+) and granulocyte (Ly-6G+ and Ly-6C+) in Atg7f/f;Lyz-Cre and wild-type mice. H, apoptosis of myeloid cells from the mouse models was assessed using Annexin V/PI staining on flow cytometer. I, flow cytometric analysis of ROS levels in Lin− cells of Atg7f/f;Vav-Cre mice or myeloid cells of Atg7f/f;Lyz-Cre mice. Data are representative or statistical results of three experiments. n ≥ 6. *, p < 0.05; **, p < 0.01.