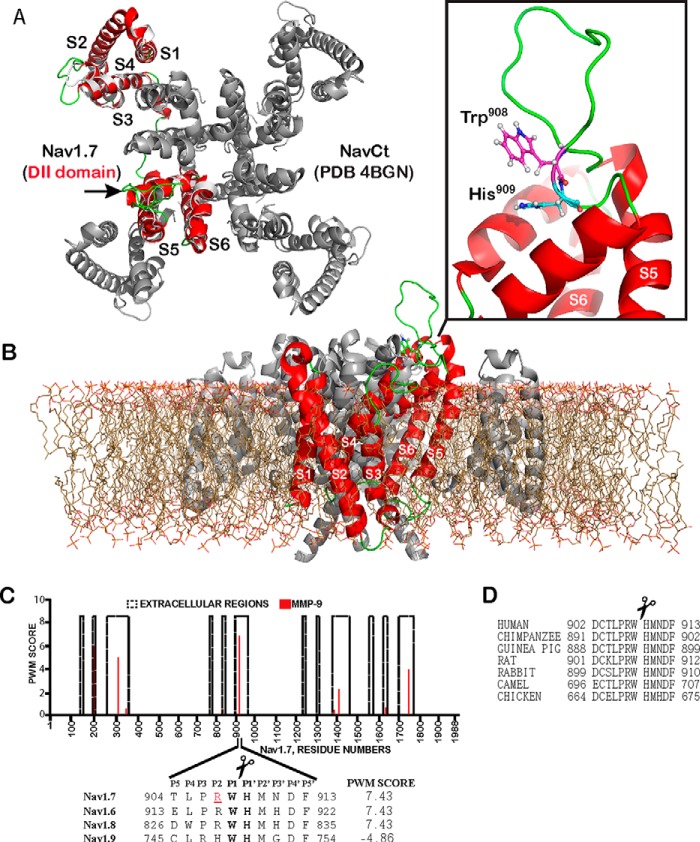
FIGURE 1.
The structure of Nav1.7 and MMP-9 cleavage site. A, superimposition of the S1–S6 transmembrane segments of the Nav1.7 DII domain (red) and the full-length C. thermarum sodium channel (NavCt, gray; PDB 4BGN). Extracellular regions are in green. An arrow points to the extracellular region connecting the S5–S6 segments. B, side view of the modeled S1–S6 transmembrane segments (red) of the domain DII with the full-length NavCt homologue (gray) immersed in a lipid bilayer. Inset, the loop (green) connecting the S5–S6 segments with the MMP-9 Trp908–His909 cleavage site (sticks). C, the PWM score of the MMP-9 cleavage sites in the Nav1.7 extracellular regions. The high PWM score suggests the presence of a potential cleavage site in the peptide sequence. Inset, the alignment of Nav1.7 with the respective regions of other sodium channels and the PWM score of the predicted MMP-9 cleavage site. The P1 and P1′ residues of the scissile bond are in bold. Arg907 (red and underlined) is mutated in CIP. D, the MMP-9 cleavage site is conserved in Nav1.7 from human, chimpanzee, guinea pig, rat, rabbit, camel, and chicken (GenBank accession numbers Q15858, H2QIX3, H0VMS3, O08562, Q28644, T0NJY4, and E1C4S2, respectively).