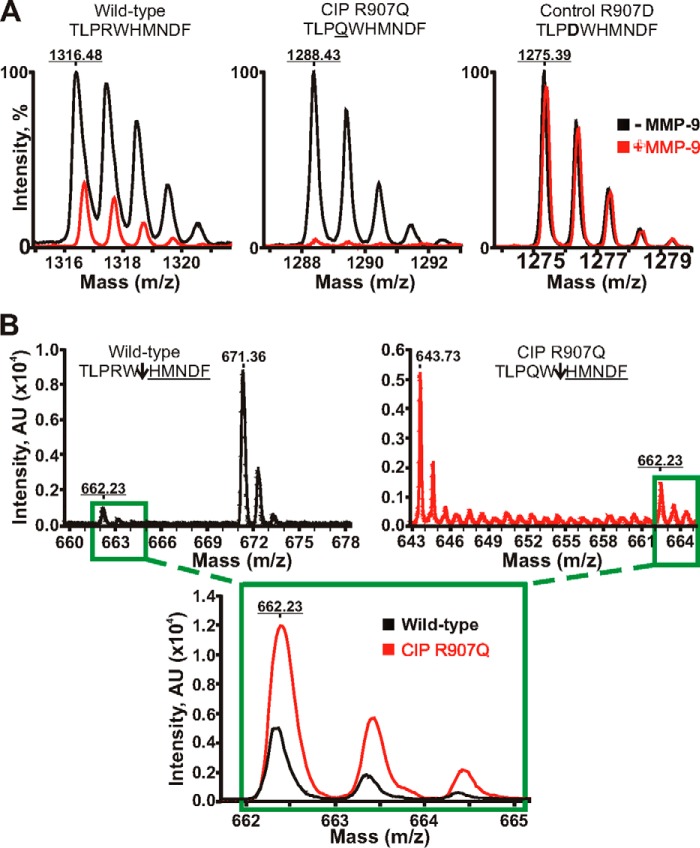
FIGURE 2.
Mass spectrometry analysis of MMP-9 proteolysis of the Nav1.7 peptides. A, MALDI monoisotopic MS spectra of the peptides. The wild-type TLPRWHMNDF (1316.48 Da), mutant TLPQWHMNDF (the CIP R907Q mutation is underlined; 1288.43 Da), and control TLPDWHMNDF (the inactivating R907D mutation is in bold; 1275.39 Da) peptides were incubated with MMP-9 at a 1:100 enzyme-substrate molar ratio. The graphs show the superimposed monoisotopic spectra of the respective intact peptides incubated with or without MMP-9 (red and black, respectively). The average mass of the intact peptides is underlined. Although the wild-type peptide and, especially, the CIP mutant were efficiently cleaved by MMP-9, the control peptide was fully resistant. B, quantification of the common HMNDF cleavage product. The wild-type peptide (black) and the R907D CIP mutant (red) were incubated with MMP-9 at a 1:250 enzyme-substrate molar ratio. Upper panels, the spectra of the TLPRW (671.36 Da), TLPQW (643.73 Da), and HMNDF (662.23 Da) cleavage products. An arrow points to the scissile bond. The molecular mass of the common HMNDF fragment is underlined. Inset, the superimposed spectra of the common HMNDF fragment generated by MMP-9 proteolysis of the wild-type and mutant CIP R907Q peptides. AU, arbitrary units.