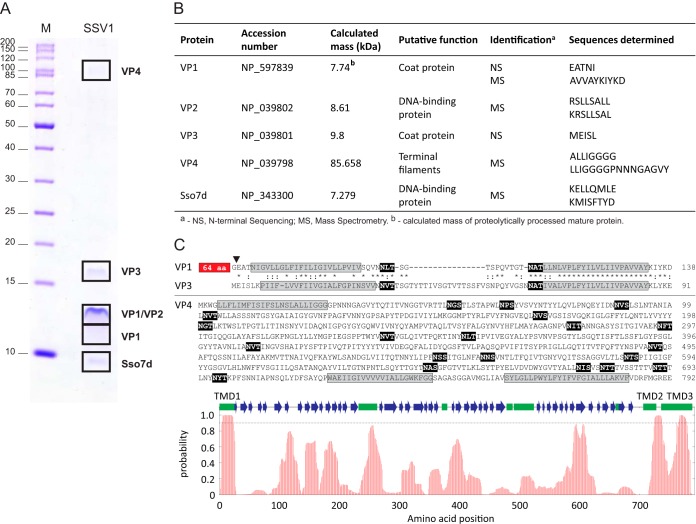
FIG 3.
Identification of the structural proteins of SSV1. (A) Protein pattern of the 2× purified SSV1 virions in a tricine-SDS-PAGE stained with Coomassie blue. M indicates the molecular mass marker. Locations of protein bands processed for proteomic analyses are depicted with black boxes, and names of proteins identified are indicated on the right. (B) Proteins identified by N-terminal sequencing (NS) and mass spectrometry (MS). The NCBI accession number, theoretical molecular masses, putative functions, and peptide sequences determined during analysis are provided. (C) Sequence analysis of the SSV1 structural proteins VP1, VP3, and VP4. Sequences of the predicted transmembrane domains are highlighted in gray, whereas the theoretical glycosylation consensus motifs (N-X-S/T) are shown on the black background. The position of proteolytic cleavage in VP1 is indicated by a black arrowhead, and the N-terminal 65-amino-acid (aa) residues not shared with VP3 are boxed. Paralogous proteins VP1 and VP3 are aligned; identical and similar amino acid positions are indicated with asterisks and colons, respectively. At the bottom of the panel is the hydrophobicity profile of VP4. The broken line indicates the 0.9 probability threshold for the prediction of the transmembrane domains (TMD1-3). Predicted secondary structure elements are shown with green boxes (α-helixes) and blue arrows (β-strands).