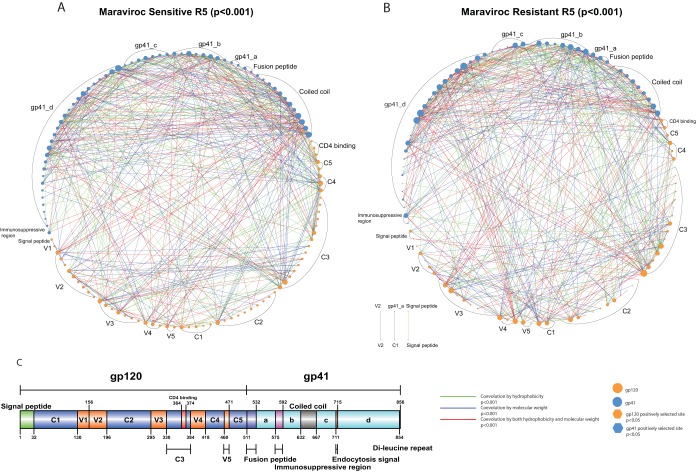
FIG 3.
Covariation networks of gp120 and gp41 in sensitive and resistant R5 viruses. (A and B) Covariation networks of HIV-1 envelope protein associated with hydrophobicity and/or molecular weight covariation for maraviroc-sensitive (A) and -resistant (B) CCR5 viruses. (C) Covarying amino acid sites are organized and labeled by protein domains in gp120 (orange circles) and gp41 (blue circles). The circle sizes indicate the relative numbers of interactions in the covariation networks. Covarying sites are linked by colored lines, which indicate the physicochemical properties of covariation (see the key below and Table 1 for a summary). Sites under positive selection are shown as hexagons. Information regarding positive selection, protein domain, and glycosylation can be found in Tables S1 and S3 in the supplemental material for covarying sites in sensitive and resistant viruses, respectively.