ABSTRACT
The lepidopteran innate immune response against RNA viruses remains poorly understood, while in other insects several studies have highlighted an essential role for the exo-RNAi pathway in combating viral infection. Here, by using deep-sequencing technology for viral small-RNA (vsRNA) assessment, we provide evidence that exo-RNAi is operative in the silkworm Bombyx mori against both persistent and pathogenic infection of B. mori cytoplasmic polyhedrosis virus (BmCPV) which is characterized by a segmented double-stranded RNA (dsRNA) genome. Further, we show that Dicer-2 predominantly targets viral dsRNA and produces 20-nucleotide (nt) vsRNAs, whereas an additional pathway is responsive to viral mRNA derived from segment 10. Importantly, vsRNA distributions, which define specific hot and cold spot profiles for each viral segment, to a considerable degree overlap between Dicer-2-related (19 to 21 nt) and Dicer-2-unrelated vsRNAs, suggesting a common origin for these profiles. We found a degenerate motif significantly enriched at the cut sites of vsRNAs of various lengths which link an unknown RNase to the origins of vsRNAs biogenesis and distribution. Accordingly, the indicated RNase activity may be an important early factor for the host's antiviral defense in Lepidoptera.
IMPORTANCE This work contributes to the elucidation of the lepidopteran antiviral response against infection of segmented double-stranded RNA (dsRNA) virus (CPV; Reoviridae) and highlights the importance of viral small-RNA (vsRNA) analysis for getting insights into host-pathogen interactions. Three vsRNA pathways are implicated in antiviral defense. For dsRNA, two pathways are proposed, either based on Dicer-2 cleavage to generate 20-nucleotide vsRNAs or based on the activity of an uncharacterized endo-RNase that cleaves the viral RNA substrate at a degenerate motif. The analysis also indicates the existence of a degradation pathway that targets the positive strand of segment 10.
INTRODUCTION
Central to the defense of insects against RNA viruses is the triggering of the so-called exogenous RNA interference pathway (exo-RNAi), by which viral double-stranded RNA (dsRNA) is cleaved intracellularly into viral small interfering RNAs (siRNAs) that in turn silence homologous transcripts (1). It is established that Dicer-2, a cytoplasmic RNase III class enzyme, recognizes and cuts successively the dsRNA template to produce small interfering RNA duplexes of ∼21 nucleotides (nt) characterized by a signature of 2-nt 3′-hydroxyl overhangs (2). These duplexes are subsequently incorporated into the effector complex RISC (RNA-induced silencing complex) via interactions with Argonaute-2, which, upon discarding one of the two strands (passenger strand), is followed by selection and cleavage of viral sequences bearing perfect complementarity to the remaining strand (guide strand) (1, 3). Dicer-2 may act upon the genome of the virus itself—as in the case of dsRNA viruses—and/or on viral replication and transcription intermediates, although structured, single-stranded RNA (ssRNA) may also be utilized (4). Interestingly, several deep-sequencing data have revealed that approximately 18- to 30-nt viral small RNAs (vsRNAs) of infected insects are not evenly distributed along the genomes but map preferentially in distinct genomic areas (hot spots) versus genomic stretches with unmapped or less-mapped vsRNAs (cold spots) (4–13). The origin of these profiles, as well as the hypothetical functional distinction between vsRNAs deriving from hot or cold spots, remains substantially elusive, although hot spots near the end of the genome have been attributed to structured regulatory viral regions (14) and an RNAi decoy-like mechanism driven by abundant vsRNAs has been proposed by another group (15). Importantly, the bulk of research on exo-RNAi biology, including deep vsRNA profiling, has been focused on Diptera (Drosophila and mosquitoes) and to a much lesser extent to Hemiptera and Hymenoptera, while studies in other orders of insects, such as Lepidoptera (moths and butterflies), are lacking considerably (16).
Bombyx mori Cypovirus 1 (B. mori cytoplasmic polyhedrosis virus or BmCPV) is a segmented dsRNA virus within the nonenveloped family of Reoviridae and a pest of sericulture that causes the cytoplasmic polyhedrosis disease in infected silkworms (17). CPV virions consist of a single-layered icosahedral capsid enclosing the tightly packaged genome. Characteristic of CPV is the existence of polyhedrin-made cubic crystals named polyhedra in which the virions are embedded for efficient protection against environmental conditions (18). The genome of BmCPV is composed of 10 segments (S1 to S10) with successively decreasing sizes ranging from 4,190 bp (S1) to 944 bp (S10), and each segment consists of a single open reading frame (ORF). In correspondence to the segments' numerals, the ORFs code for the structural proteins VP1, VP2, VP3, VP4, VP6, and VP7, including an RNA-dependent RNA polymerase (RdRp) transcribed from S2; the nonstructural proteins NSP5, NSP8, and NSP9; and polyhedrin, which is transcribed from S10 (see reference 19 and references therein). BmCPV infection occurs via the fecal-oral route, and the disease is mainly restricted to the larval midgut. Accordingly, after the consumption of contaminated food by the larvae, the polyhedra dissolve inside the highly alkaline environment of the lepidopteran midgut, releasing virions which adhere to and penetrate the plasma membrane of microvilli and finally settle in the cytoplasm of the columnar epithelial cells (20). RdRp-mediated synthesis of positive strands (viral mRNAs) initiates inside the capsid, and positive-strand RNAs are further capped and released in the cytoplasm for protein translation. Finally, a population of positive-strand RNAs incorporates into newly formed capsids, where the negative RNA strands of the genome are synthesized, and the virions embed into polyhedra, which are in turn released, along with the feces, into the environment after host cell lysis (20, 21).
In a previous work, we exploited a laboratory silkworm strain (Daizo) that was persistently infected with BmCPV in the absence of observable signs of pathogenicity and analyzed the transcriptome of midgut cells after acute, pathogenic infection with the same virus (22). Deep-sequencing analysis and comparison with data from literature revealed that a preexisting (low-level) persistent infection does not influence profoundly the antiviral response against the pathogenic infection. Importantly, the data also suggested an important role for exo-RNAi in both persistent and pathogenic infections, as demonstrated by the detection of 20-nt vsRNAs in both states of infection and an upregulation of expression of core RNAi genes during acute infection (22). In the present study, we analyze in detail the Illumina-based deep-sequencing data of vsRNAs of persistent and pathogenic BmCPV infections, and this is, to our knowledge, the first study on deep vsRNA profiling of an RNA virus in Lepidoptera.
MATERIALS AND METHODS
Silkworm rearing, infection of BmCPV, RNA extraction, and deep sequencing of RNA samples.
Silkworm rearing, infection of larvae with BmCPV, RNA extraction, and deep sequencing of RNA samples from the midguts of persistently and pathogenically infected larvae are described elsewhere (22). In addition, the reads of the 50-nt libraries that map to the BmCPV genome (i.e., reads derived from mRNA, genomic RNA, and replication intermediates) were used to calculate the RPKM (Reads Per Kilobase per Million mapped reads) values of the positive and negative strands of each of the viral segments. Distribution graphs of the lengths of small RNAs and distribution profiles of vsRNAs on the viral segments were produced in R (23) by use of the viRome package (http://www.ark-genomics.org/bioinformatics/virome). The sequences from the “transcriptome” and small-RNA libraries of all four samples were submitted to the European Nucleotide Archive (accession number PRJEB7502 [http://www.ebi.ac.uk/ena]).
Dicer-2 processing signature.
For the evaluation of Dicer-2 processing signature, we analyzed the 17- to 23-nt vsRNAs derived from the pathogenically infected fourth-instar larvae as Dicer-2 activity is enhanced in this larval stage. The reads' matrices of the individual 17- to 23-nt classes consisted of a table, where the two columns corresponded to the positive or the negative viral strands, and the rows represented the genomic position of vsRNAs belonging to the different size classes. The correlation between the positive- and negative-strand data columns was estimated with the Pearson correlation coefficient (r). Specifically, in complete alignment of the matrices (position 0) the reads of the negative and positive strands correspond to identical genomic positions. Next, the negative-strand read column was shifted (upstream or downstream with respect to the positive strand) by successive steps of 1 nt, and r was recalculated for each such shift. The statistical significance (P < 0.05) was estimated with one-tailed tests.
Identification of a degenerate motif at the cleavage sites of the vsRNAs.
For the identification of the degenerate motif, we analyzed the 20- to 29-nt vsRNAs derived from the pathogenically infected second-instar larvae as 22- to 29-nt vsRNAs are more abundant in this larval stage. Specifically, all 20- to 29-nt vsRNA sequences with common 5′ or 3′ ends were selected and then analyzed by taking into account 10 nt of the flanking genomic sequences of their 5′ or 3′ ends, respectively. Both positive- and negative-strand-derived vsRNAs were evaluated. The four sequence collections were analyzed with WebLogo 3 program (http://weblogo.threeplusone.com/create.cgi) (24). The statistical significance (P < 0.05) of enriched or depleted bases of the created logos was evaluated with one-proportion Z-tests as previously described (25). For the delineation of the degenerate motif of DVNV̂DC at the cut site (^) of vsRNAs, only the positions with statistically significantly enriched or depleted bases in the logos of all four sequence collections were selected.
To correlate the presence of vsRNAs with the degenerate motif, all sites of the degenerative motif were first determined in the BmCPV genome sequence (positive and negative strands were separately assessed). The 5′ and 3′ ends of the vsRNA reads were subsequently correlated with the predicted cleavage sites. First, all sites of the DVNV̂DC motif in BmCPV genome were assayed for the construction of a “(1,0)” binary matrix, designated as a genome motif matrix, indicating the existence—“(1)”—or nonexistence—“(0)”—of a V̂ in a particular genomic position. Thus, the genome motif matrix indicates the degenerate motifs whose V̂ are positioned at the 5′ ends of vsRNAs of all 20- to 29-nt classes. Subsequently, 10 class-specific binary matrices, designated as “n”-nt motif matrices (n = 20 to 29), were each respectively constructed by the additional input of value 1 in the x-n positions of the genome motif matrix, where “x” denotes all positions of the genome motif matrix whose values are equal to 1. Thus, an “n”-nt motif matrix, in addition to the aforementioned 5′ V̂ sites, indicates the degenerate motifs whose N̂ are positioned in the 3′ ends of the x-n positioned vsRNAs. Next, the deep-sequencing vsRNA data matrices of each of the 20- to 29-nt classes were transformed into “(1,0)” binary matrices, designated “n-nt reads” matrices, indicating the existence (1) or nonexistence (0) of more than 1 reads of the vsRNA mapped to the various genomic positions. Single-read vsRNAs were excluded from the analysis since they were found to produce additional background. Finally, the correlation of the n-nt reads' matrices and the corresponding n-nt motif matrices were evaluated with the Pearson correlation coefficient (r). r values were reassessed for single nucleotide shifts of one of the two matrices in comparison, corresponding to the shifted degenerate motif as internal controls. Statistical significance (P < 0.05) was estimated with one-tailed tests.
Functional assay for knockdown of polyhedrin mRNA by artificial siRNAs (asiRNAs) and segment 10 hairpin RNAs.
The BmCPV polyhedrin gene was RT-PCR amplified from segment 10 with the primers 5′-AATTGGATCCCAACATGGCAGACGTAGCAGGAACAAG-3′ (forward) and 5′-CCGGATCCCTGACGGTTACTCAGAGCT-3′ (reverse), using 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min, and then cloned into the BamHI site of the pEA-MycHis expression vector (26), thus generating the expression plasmid pEA-polyhedrin-MycHis. For calibration of the transfection efficiency, pEA-polyhedrin-MycHis was cotransfected with an expression vector that encodes the capsid protein of Flock House virus (FHV) fused to a C-terminal MycHis tag (pEA-FHV-CP-MycHis). The capsid protein of FHV was amplified from cDNA of persistently infected Hi5 cells (27) using 5′-AATTGGATCCCAACATGGTCAACAACAGCAGACCAAAAC-3′ (forward) and 5′-AATTGGATCCGTCTAAAATCCAAAACCTTCAAATAG-3′ (reverse) as primers, and the PCR fragment was subcloned into the BamHI site of the pEA-MycHis vector as described for BmCPV polyhedrin.
The designed asiRNAs (see Table 2) were supplied by Eurogentec (Belgium). The secondary structure of the positive-strand segment 10 was predicted with RNAfold (28), and the identified hairpin (s10-H) spanning positions 707 to 788 was evaluated with MiPred (29). For hairpin RNA synthesis of segment 10 hairpin (s10-H) and of segment 10 mutated hairpin (s10-mutH), the oligonucleotide pair of 5′-AATGGTACCTCACTACGTTTCACCGAATGCTTACCCATATCTCGATATTAACAATCACAGTTATGGAGTAGCTTTGAGTAACCGTCAGTGAGAGCTCTAA-3′ and 5′-TTAGAGCTCTCACTGACGGTTACTCAAAGCTACTCCATAACTGTGATTGTTAATATCGAGATATGGGTAAGCATTCGGTGAAACGTAGTGAGGTACCATT-3′ (for s10-H) and the oligonucleotide pair of 5′-AATGGTACCACTCTTCCTATCTCGGATTGCTTTCCGATTTGTCCAAATTAACATTGACACTAATCGAGAAGCATTCAGAATCGGACAGAGTGAGCTCTAA-3′ and 5′-TTAGAGCTCACTCTGTCCGATTCTGAATGCTTCTCGATTAGTGTCAATGTTAATTTGGACAAATCGGAAAGCAATCCGAGATAGGAAGAGTGGTACCATT-3′ (for s10-mutH; mutated sites are indicated with underlined bases of the sense oligonucleotide) were incubated in annealing buffer (92°C for 5 min and then left to gradually cool down), and the produced dsDNA fragments were subsequently cloned into the EcoRV-site of pLitmus-38i vector (New England BioLabs), generating the pLitmus-s10-H and pLitmus-s10-mutH vectors, respectively. After verification by sequencing, the vectors were linearized with BamHI and then used as the templates for in vitro transcription to synthesize hairpin RNA by T7 RNA polymerase (Thermo Scientific). The reactions were subjected to DNase I treatment and purification (phenol-chloroform extraction and ethanol precipitation). The hairpin RNA concentration was estimated by comparison to known amounts of Lambda DNA marker (HincII/HindIII digestion) electrophoresed in 2% agarose gels and visualized with ethidium bromide staining. Transfection of Hi5 cells was carried out according to established protocols (30) using Escort IV (Sigma) as transfection reagent. For polyhedrin silencing studies, the cells were cotransfected with 0.9 μg of pEA-polyhedrin-MycHis/ml, 0.9 μg of pEA-FHV-CP-ΜycHis (transfection normalization vector)/ml, 0.3 μg of pBmIE1 helper plasmid/ml, and either 20 nM asiRNAs (asiRNA-mediated silencing) or 200 nM hairpin RNA (hairpin-mediated silencing). The cells were harvested at 3 days posttransfection, and the total cell extracts were collected by freeze-thawing as described previously (31). The experiments were performed in triplicates.
TABLE 2.
asiRNA pairs that mapped to hot, moderate, and cold spots of S10a
| asiRNA | Sequence (5′-3′) | Position | No. of reads |
|
|---|---|---|---|---|
| 2inf | 4inf | |||
| h.1p | CAGACGUAGCAGGAACAAGT | 46 | ||
| h.1n | UUGUUCCUGCUACGUCUGCC | 44 | 81 | 207 |
| h.2p | GGACGCGAACAAAGACUATT | 81 | ||
| h.2n | UAGUCUUUGUUCGCGUCCGC | 79 | 269 | 478 |
| h.3p | CUAUUCAACAGCGAACAATA | 96 | ||
| h.3n | UUGUUCGCUGUUGAAUAGTC | 94 | 67 | 140 |
| h.4p | GGACAACACAAUGACUCUTA | 261 | ||
| h.4n | AGAGUCAUUGUGUUGUCCCT | 259 | 42 | 158 |
| m.1p | CAUACUACUCAGACGGGUCA | 163 | ||
| m.1n | ACCCGUCUGAGUAGUAUGCG | 161 | 18 | 53 |
| m.2p | CAAUGACUCUUACGAUGAAT | 269 | ||
| m.2n | UCAUCGUAAGAGUCAUUGTG | 267 | 3 | 36 |
| m.3p | CGCAAGAUUAAGAGAGACGG | 492 | ||
| m.3n | GUCUCUCUUAAUCUUGCGGT | 490 | 14 | 34 |
| m.4p | GUAAUGGACGUCUUCCAATC | 630 | ||
| m.4n | UUGGAAGACGUCCAUUACGT | 628 | 5 | 28 |
| c.1p | CGAGGUGAACCAAGGCAUCT | 290 | ||
| c.1n | AUGCCUUGGUUCACCUCGTA | 288 | 0 | 0 |
| c.2p | GGCGGUGACGCUCGUAAATA | 321 | ||
| c.2n | UUUACGAGCGUCACCGCCGT | 319 | 0 | 0 |
| c.3p | GAACUACCCCAUGAAUAUAA | 540 | ||
| c.3n | AUAUUCAUGGGGUAGUUCAA | 538 | 0 | 0 |
| c.4p | CGUAAUGGACGUCUUCCAAT | 629 | ||
| c.4n | UGGAAGACGUCCAUUACGTT | 627 | 1 | 9 |
| c.4.21a.p | CGUAAUGGACGUCUUCCAATC | 629 | ||
| c.4.21a.n | UUGGAAGACGUCCAUUACGTT | 627 | 0 | 0 |
| c.4.21b.p | GUAAUGGACGUCUUCCAAUCA | 630 | ||
| c.4.21b.n | AUUGGAAGACGUCCAUUACGT | 628 | 0 | 0 |
| nsp | CAAACGACCCAUACAUGACA | |||
| nsn | UCAUGUAUGGGUCGUUUGAA | |||
The genomic positions are shown in the position column. Deep-sequencing reads of the corresponding negative-strand-derived vsRNAs (which target polyhedrin mRNA) are shown in columns 4 and 5 for pathogenically infected second- and fourth-instar larval samples (2inf and 4inf), respectively. Nucleotides in 3′ overhangs are deoxynucleotides and are therefore indicated as “T” instead of “U.” nsp and nsn, a nonspecific asiRNA pair. The “h.,” “m.,” and “c.” prefixes refer to hot, moderate, and cold spots, respectively.
The protein samples of transfected cells were subjected to Western blot analysis essentially as described previously (32). The samples were electrophoresed in 12% polyacrylamide gels, the separated proteins were subsequently transferred to nitrocellulose blotting membranes (Amersham), and the membranes were blocked with 10% nonfat milk. For the detection of the MycHis-tagged proteins, anti-Myc (Cell Signaling) at 1:1,000 as the primary antibody and horseradish peroxidase-conjugated anti-mouse (Millipore) at 1:2,000 as the secondary antibody were used. Densitometry analysis was performed using ImageJ software (33).
RESULTS
Abundance of viral segments and vsRNAs differs significantly between conditions of persistent and pathogenic infection.
For the analysis of BmCPV infection by deep sequencing, two types of libraries were constructed (22). The first type of library represents the protein coding transcripts of the infected cells, and it also contains the reads originating from the positive and negative strands of the dsRNA viral genome segments. The second type of library represents the small RNAs, such as the miRNAs, of the infected cells, and it also contains the reads of the vsRNAs. As in a previous study (22), samples from persistently and pathogenically infected second-instar larvae were designated “2c” and “2inf,” respectively, whereas the corresponding samples from the fourth instar were designated “4c” and “4inf.”
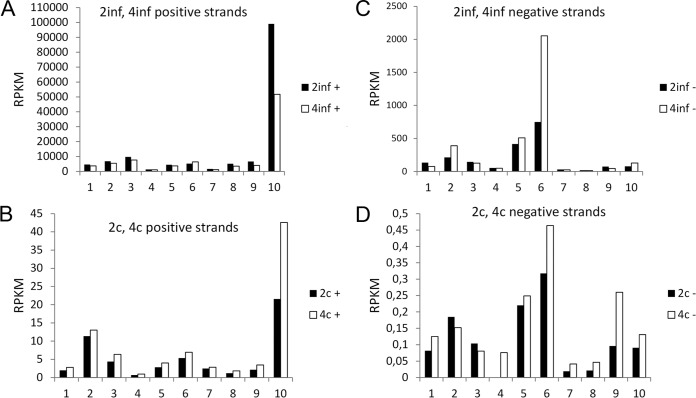
Regarding the first type of library, RPKM values of positive and negative strands of individual viral segments in pathogenically infected 2inf samples are on average ∼2,220- and ∼1,390-fold higher, respectively, than in persistently infected 2c samples. Similarly, the aforementioned RPKM values are on average ∼1,600- and ∼1,390-fold higher in 4inf samples compared to 4c samples. Collectively, the data indicate very high differences in viral loads between the two types of infection and in particular suggest very low viral loads in persistent, nonpathogenic infections.
In both types of infection the RPKM values in the first type of library for all 10 BmCPV segments demonstrate a positive-strand bias which could be explained by the additional populations of free positive strands that constitute the viral transcripts (Fig. 1, compare the scale of RPKM in the graphs for the plus and minus strands in both types of infection). Interestingly, RPKM values of the positive strand of segment 10 (which encodes the polyhedrin gene) in pathogenically infected larvae are markedly higher than of the rest of the segments (Fig. 1A), while a milder enrichment is also observed for the persistently infected larvae (Fig. 1C). Given that the RPKM values of (−) RNAs of segment 10 are not proportionally increased compared to the other segments (Fig. 1), and thus the possibility of an enrichment of the genomic dsRNA of segment 10 is excluded, we conclude that the observed peaks of RPKM values of positive-strand RNAs of segment 10 indicate an overexpression of polyhedrin transcript in both pathogenically and persistently infected silkworms. A similar analysis on the small-RNA libraries revealed that the persistently infected second- and fourth-instar larvae (2c and 4c, respectively) also demonstrate a markedly lower abundance of vsRNAs in individual viral segments compared to animals with acute, pathogenic infection (2inf and 4inf) (Table 1). Accordingly, the proportion of 16- to 37-nt vsRNA reads mapped to BmCPV in the total of Bombyx-filtered reads is <0.1% in the persistently infected larvae, whereas the same proportion reaches 49% in the 4inf larvae (Table 1).
FIG 1.
Reads per kilobase per million mapped reads (RPKM) of individual BmCPV segments' positive (+) (A and C) and negative (−) (B and D) strands based on the 2inf and 4inf 50-nt (“transcriptome”) libraries (see Materials and Methods and Results). In the horizontal axes, numerals of the BmCPV segments (numbered 1 to 10) are indicated. 2c and 4c refer to persistently infected samples of second- and fourth-instar larvae (C and D), while 2inf and 4inf refer to pathogenically infected samples of the same developmental stages (A and B).
TABLE 1.
Illumina read counts for 16- to 37-nt RNAs that are not mapped to Bombyx mori genome (Bombyx filtered) but are mapped to BmCPV genome in various samplesa
| Sample | No. of Bombyx mori-filtered counts | No. (%) mapped to BmCPV |
|---|---|---|
| 2c | 1,609,207 | 748 (0.05) |
| 4c | 1,902,001 | 1,179 (0.06) |
| 2inf | 4,303,833 | 1,614,171 (37.51) |
| 4inf | 5,863,649 | 2,872,716 (48.99) |
“2c” and “4c” refer to midgut samples that were persistently, nonpathogenically infected with BmCPV at the second- and fourth-instar larval stages, respectively. “2inf” and “4inf” refer to midgut samples that were pathogenically infected at the same larval stages (21). In parentheses are the percentages of BmCPV-mapped vsRNA reads in the Bombyx mori-filtered 16- to 37-nt reads.
Distribution profiles of the lengths of the vsRNAs in infected larvae.
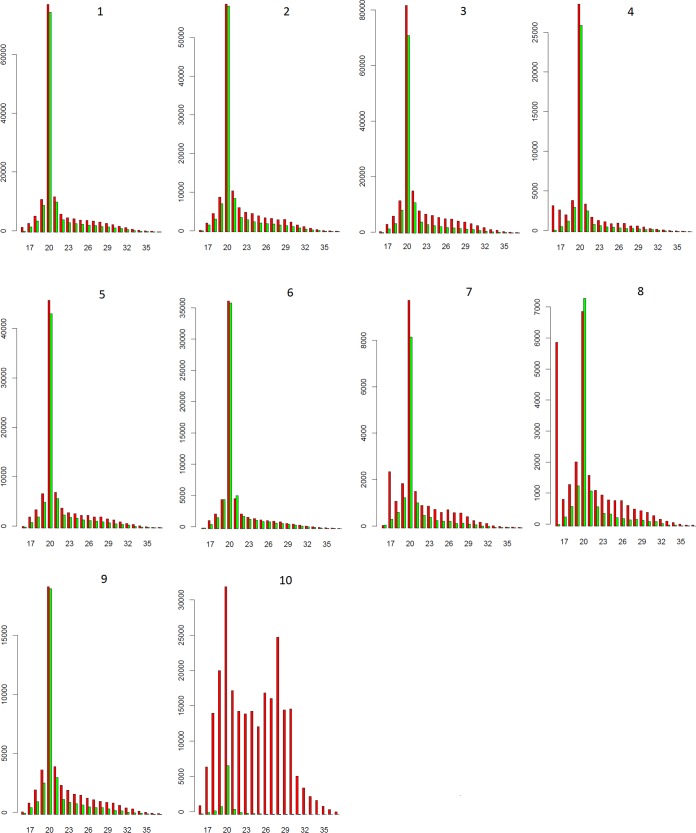
Analysis of vsRNA size revealed that the 20-nt class is the predominant class among the assessed 16- to 37-nt vsRNAs for all viral segments (Fig. 2 shows the distribution graph for the 2inf sample), whereas the flanking classes of 19 and 21 nt are, in general, also distinguished from the rest. vsRNA reads mapping to positive and negative strands are comparable for the 19- to 21-nt classes of the various segments, with the exception of vsRNAs originating from S10 for which a strong positive-strand bias is observed (see also further below). Thus, in contrast to the abundance of the strands of the viral segments where a clear bias of the positive strand is observed (Fig. 1), the 19- to 21-nt vsRNAs map in almost equal abundance to plus and minus strands (Fig. 2). This likely reflects their main origin from dsRNA structures (e.g., genomes and replication intermediates), as opposed to structured regions in ssRNA strands (transcripts).
FIG 2.
Sequence length distribution of BmCPV-derived 16- to 37-nt vsRNAs from pathogenically infected second-instar larval midguts. Individual viral segments (numbered 1 to 10) are depicted. Profiles corresponding to the positive and negative strands are represented by red and green bars, respectively. Vertical axes show the number of vsRNA reads.
Regarding most of the other vsRNA classes (i.e., not 20 nt), a mild bias toward the positive strand is observed for most segments (Fig. 2). Further analysis (below) indicates that the vsRNAs of size classes different from 19 to 21 nt do not carry the signature of Dicer-2 cleavage and are proposed to be generated by a different mechanism. The bias toward the positive strand is especially pronounced for S10 and encompasses vsRNAs of all different sizes, including 19 to 21 nt, for which a Dicer-2 signature exists. More specifically, an additional peak of vsRNAs mapping to the positive strand of S10 is observed at 28 nt, which is almost of similar abundance as the 20-nt class (Fig. 2).
Also, the 16- and 17-nt vsRNAs originating from S4, the 17-nt vsRNAs originating from S7, and the 16-nt vsRNAs originating from S8 display a strong bias toward the positive strand. The vsRNAs 16 to 17 nt in length with positive-strand bias map at the ends of these genome segments, implying that these particular ends are, for some as-yet-unknown reason, exceptionally labile and/or exposed to RNases (see also the Discussion).
Mapping of vsRNAs on viral dsRNA genome segments.
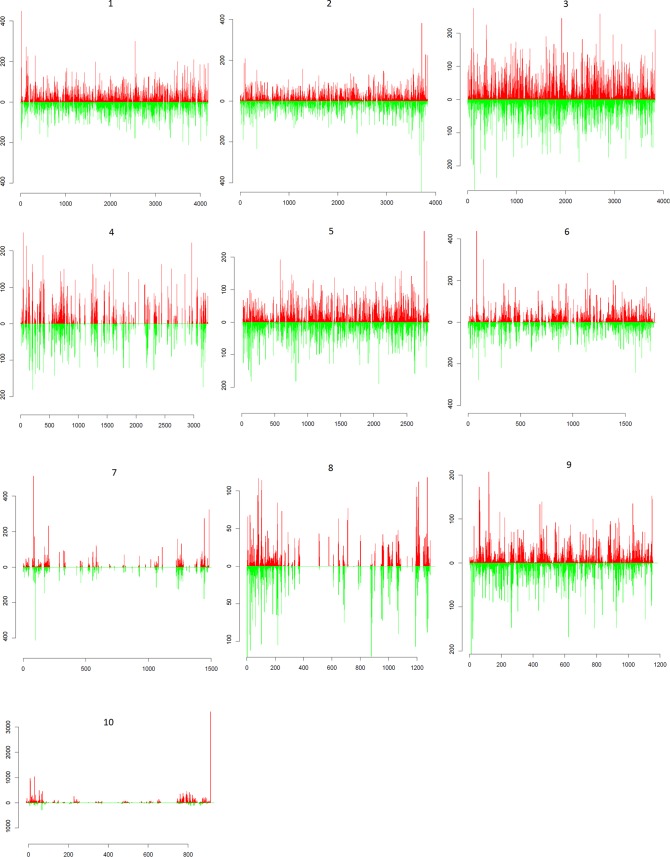
Genome coverage of the 20-nt vsRNA class in pathogenically infected larvae ranges between 28% for S7 of the 2inf samples to 93% for S9 of the 4inf samples. From the analysis, it is apparent that vsRNAs of pathogenically infected silkworms are not mapped evenly throughout the BmCPV genome (Fig. 3). In particular, the distributions of vsRNAs reveal distinct hot and cold spot patterns, clearly observable for segments 4, 7, 8, and 10 (Fig. 3), which have the lowest overall segment-wide vsRNA coverage. Importantly, these hot and cold spot profiles are very similar for the 2inf and 4inf samples (see Fig. 7 for representative data), suggesting a nonrandom origin of the respective vsRNAs distributions.
FIG 3.
Distribution profile of BmCPV-derived 20-nt vsRNAs from midgut samples of pathogenically infected second-instar larvae (2inf). Horizontal axes depict the nucleotide sequence positions of 1 to 10 viral segments in 5′→3′ direction for the positive strand and in a 3′→5′ direction for the negative strand. RNAs mapped to the positive and negative strands are represented by red and green bars, respectively. Vertical axes show the number of vsRNA reads.
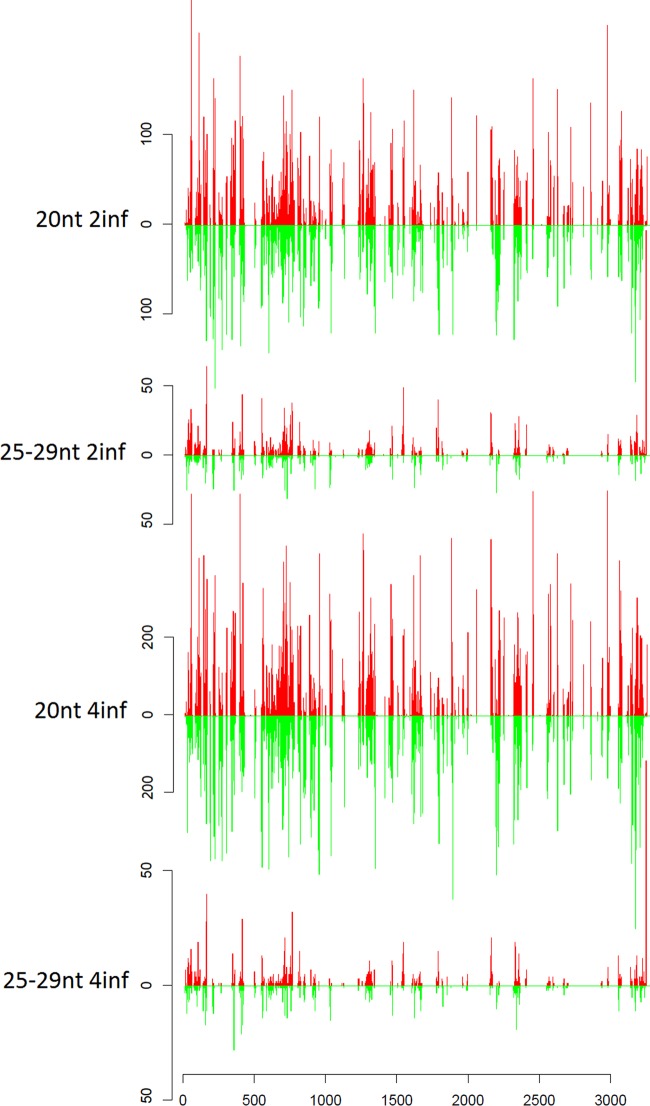
FIG 7.
Distribution profiles of the 20-nt and 25- to 29-nt vsRNA classes that map to S4 of BmCPV from the 2inf and 4inf samples. Horizontal axes depict the nucleotide sequence position in a 5′→3′ direction for the positive strand and the 3′→5′ direction for the negative strand. The abundances of vsRNAs mapping to the positive and negative strands are represented by red and green bars, respectively. Vertical axes show the number of vsRNA reads.
Following the finding that vsRNAs are markedly decreased in the persistently infected silkworms, we sought also to investigate the distributions of the vsRNAs derived from the samples of persistently infected larvae (2c and 4c), particularly in comparison to the respective profiles found in pathogenically infected larvae (2inf and 4inf). However, vsRNAs from 2c and 4c samples are in general refractory to statistical evaluation due to their low abundance, e.g., not all of the 16- to 37-nt classes are represented in the distributional maps of the various viral segments. Nevertheless, vsRNA numbers also peak at 20 nt for most segments and the whole genome, indicating that exo-RNAi is operative in both pathogenic and persistent infections (22). Since persistent and pathogenic infections are clearly distinct with respect to pathology, an intriguing question is whether the vsRNA profiles are also functionally distinct between the two types of infections. The analysis, however, revealed that practically all of the vsRNAs of 2c and 4c samples are also found in the samples of the pathogenically infected larvae. Moreover, very few vsRNAs are common between 2c and 4c samples. To illustrate this, four common vsRNAs of 20 nt were detected in 164 and 240 unique vsRNAs that mapped to the negative strand, and six common vsRNAs were detected in 156 and 231 unique vsRNAs that mapped to the positive strand in samples of persistently infected larvae at the two different stages. Thus, in contrast to the 2inf and 4inf vsRNA profiles, the 2c and 4c vsRNA profiles are nonoverlapping. We therefore propose that vsRNAs of persistently infected samples correspond to a small, random fraction of the vsRNAs produced in pathogenic infections.
Dicer's signature in the production of 20-nt vsRNAs from dsRNA.
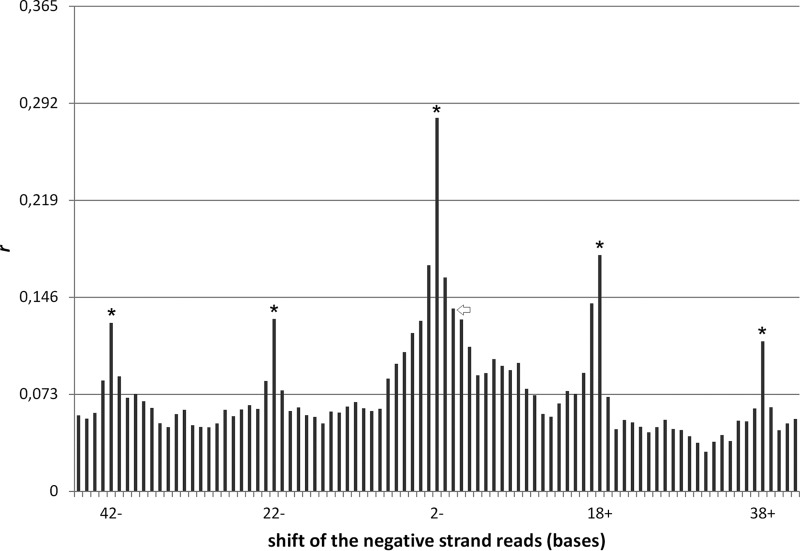
For evaluating a possible correlation between positive- and negative-strand 20-nt vsRNAs, a statistical analysis using the Pearson correlation coefficient (r) was performed. The r value assessing whole-genome positive- and negative-strand reads of identical positions, which corresponds to RNA duplexes with no overhangs, is indicated by an arrowhead in Fig. 4 (position 0). Subsequently, the negative-strand data column of the deep-sequencing reads was shifted by successive steps of 1 nt in an upstream (negative) or downstream (positive) direction with respect to the positive-strand data column, and r was recalculated for each such shift. The highest r value was calculated for position −2 (the highest peak marked with an asterisk in Fig. 4) which corresponds to RNA duplexes with 2-nt 3′ overhangs characteristic of Dicer-2 processing. Another significant finding is the recurrence of r value peaks every 20 nt counting from position −2 irrespective of the strand's relocation direction (also marked with asterisks in Fig. 4). These peaks demonstrate that the densities of 20-nt reads of the positive strands are correlated to densities of the negative-strands' reads precisely at integer multiples of 20. Taken together, the data suggest that Dicer-2 processes viral dsRNA and cuts successively every 20 nt, thus producing the abundant 20-nt vsRNAs class. Other vsRNA classes (i.e., 17 to 19 nt and 21 to 23 nt) were also assessed, but neither demonstrated statistically significant repetitive r values, whereas only the 19- and 21-nt classes showed statistically significant r values around position −2. Interestingly, when we tested a 20-nt artificial small interfering RNA (asiRNA) mapped to the positive strand of segment 10 and compared its activity with two 21-nt asiRNAs that are extended by one base each at the 5′ or 3′ end of the former, it was observed that all three asiRNAs efficiently silence their cotransfected transgene target (data not shown). Thus, among the relatively infrequent 19- and 21-nt vsRNAs produced by Dicer-2 at least the latter class appears to have silencing activity similar to that of the 20-nt class in B. mori.
FIG 4.
Estimation of the correlation of 20-nt reads mapped to the positive and negative strands of BmCPV dsRNA genome (fourth-instar larvae). Negative (−) and positive (+) positions of the horizontal axis indicate the relative positions of the negative-strand reads upstream and downstream of the positive-strand reads, respectively. Correlation peaks are observed at the relative positions −42, −22, −2, +18, and +38 and correspond to annealing of positive and negative strands with a 2-nt 3′ overhang with repetition every 20 nt, a finding consistent with cleavage by the Dicer-2 enzyme. The arrow indicates the bar at position 0, corresponding to complete alignment between negative and positive strands in the absence of positional shift. Statistical significance (P < 0.05) for a positive correlation of a one-tailed test is given for r > 0.073 and marked by asterisks for the peaks that indicate Dicer-2 processing. r, Pearson correlation coefficient.
Both hot and cold spot asiRNAs efficiently silence their target.
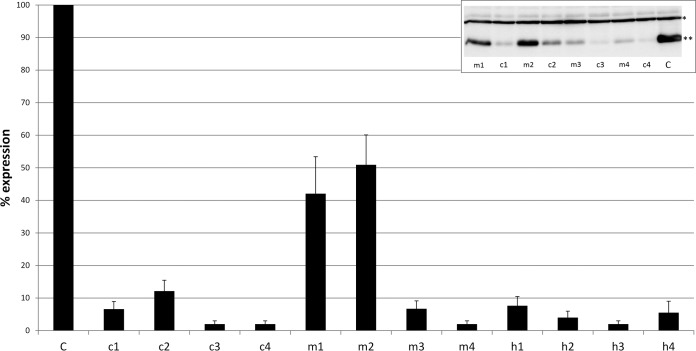
To elucidate whether 20-nt vsRNAs corresponding to different topologies exhibit different efficacies in silencing their target, we exploited a collection of asiRNAs targeting the positive strand of segment 10 and evaluated the silencing of a MycHis-tagged polyhedrin gene via Western blot densitometry in cotransfection experiments. The asiRNAs that were tested map to either hot or cold spots in the S10 distribution profiles of pathogenically infected larvae, although intermediate moderate spot asiRNAs were also included (see Materials and Methods and Table 2). The results indicated that asiRNAs from all three classes potently silence the MycHis-tagged polyhedrin target, reducing the protein's level by >80%, with the exceptions of moderate spot 1 and 2 (m1 and m2) asiRNAs, which are less effective (Fig. 5). Therefore, no clear evidence was found to support the initial hypothesis of differential silencing efficacies between hot and cold spot-derived vsRNAs.
FIG 5.
Artificial siRNA (asiRNA)-mediated knockdown of BmCPV polyhedrin presented as the percent expression of the control (C, nonspecific asiRNA). c1 to c4, m1 to m4, and h1 to h4 asiRNA map to the cold, moderate, and hot spots of BmCPV segment 10, respectively. A representative Western blot showing knockdown of polyhedrin by moderate and cold spot asiRNA is shown in the inset image. Specific proteins detected by Myc antibody were FHV-cp1-MycHis (*; 45 kDa; transfection normalization protein) and BmCPV-polyhedrin-MycHis (**, 30 kDa).
Structured ssRNA as an additional source for vsRNA production.
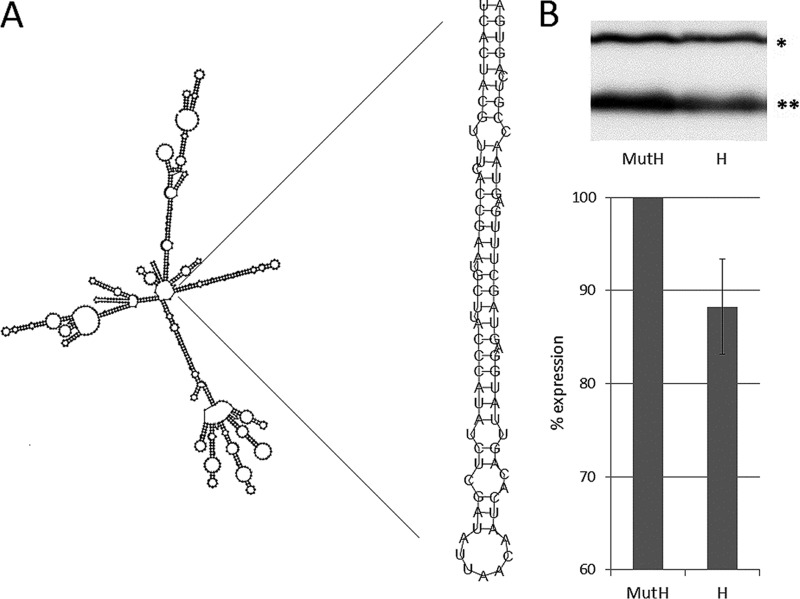
The positive-strand bias observed in the sequence length distribution of vsRNAs on S10 (Fig. 2 and 3) indicates that the polyhedrin transcript can function as an additional source for the production of vsRNAs. Because the (+)-vsRNAs show a very distinct distribution in length from 17 to 30 nt (Fig. 2) and because of the extensive overlap between the 2inf and 4inf samples, it is unlikely that they are random degradation products of the positive strand of S10. It has been shown, for example, that Dicer-2 may also process structured ssRNA of viral intramolecular stem-loops (4). Furthermore, Drosha, another RNase III class enzyme which processes primary micro-RNAs (pri-miRNAs) stem-loops in the initial, nuclear step of miRNA biogenesis, was reported to shift to the cytoplasm upon viral infection and process virus-derived cytoplasmic primary miRNAs (c-pri-miRNAs) (34). Given the high abundance of (+)-vsRNAs originating from S10, we first sought to determine whether stem-loops are found in the predicted secondary structure of the positive strand of S10. Using RNAfold, several stem-loop structures were found, among them an extended hairpin 82 nt in length (Fig. 6), which is further predicted to form a miRNA precursor (pre-miRNA) by the MiPred program. This hairpin stretches along a hot spot of the positive strand of segment 10, which is most pronounced in the 2inf sample. Next, we tested whether artificially produced hairpins of the respective sequence could affect the level of tagged polyhedrin expression in cotransfection experiments and found a modest (13%) but repeatable reduction of the protein's level compared to a mutated hairpin (Fig. 6). Thus, our data demonstrate that pre-miRNA-like hairpins from structured polyhedrin mRNA may provide an additional template for vsRNA biogenesis.
FIG 6.
Structured regions in transcripts of S10 can function as an additional source of vsRNAs. (A) Predicted secondary structure of the positive strand of S10 (or the polyhedrin transcript) using RNAfold. The detail depicts the 82-nt stem-loop that overlaps with a hot spot for (+)-vsRNA production. (B) Hairpin (H)-mediated knockdown of BmCPV polyhedrin expression in Western blot compared to mutated hairpin (MutH). Hi5 cells were cotransfected with MycHis-tagged BmCPV-polyhedrin and Myc-tagged FHV-cp1 (for normalization) expression constructs, together with either H RNA corresponding to the predicted 82-nt stem-loop or MutH RNA. A representative Western blot showing knockdown of polyhedrin by H RNA is shown at the top of the panel. Specific proteins detected by Myc antibody were FHV-cp1-MycHis (*; 45 kDa; transfection normalization protein) and BmCPV-polyhedrin-MycHis (**, 30 kDa).
Indications for an endonucleolytic activity prior to Dicer-2 processing.
As already stated above, the hot and cold spot maps of the various vsRNA classes highly overlap between the two developmental stages of pathogenic infections. To illustrate this, we provide as an example the 20-nt and 25- to 29-nt vsRNA distribution profiles of segment 4 for the second (2inf)- and fourth (4inf)-instar larvae samples in a vertical alignment (Fig. 7). The extensive overlap of the vsRNA distributions for 20-nt vsRNAs and for 25- to 29-nt vsRNAs between the 2inf and 4inf samples is obvious, although the latter are less abundant. Since only the 19- to 21-nt vsRNAs classes were related to Dicer-2 processing, we therefore assumed that the reproducible, nonrandom distribution of the >21-nt vsRNAs classes is possibly connected to an unknown endonucleolytic RNase activity.
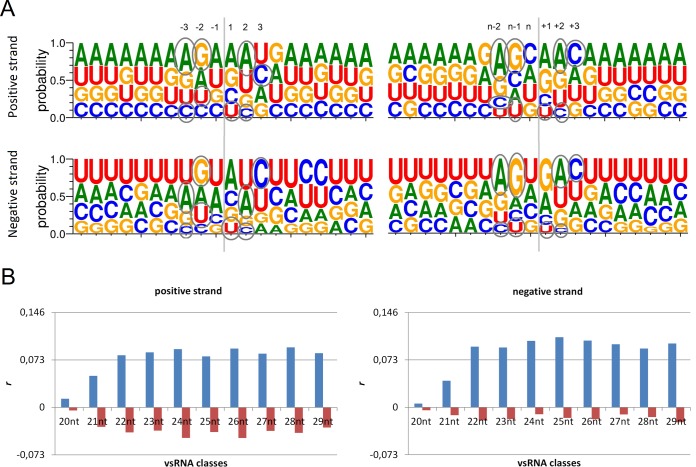
Given the similarity of hot and cold spot maps, we sought to determine whether a relation between these similarly positioned vsRNAs of the various classes exists with respect to the viral genomic sequence. To examine this possibility, we selected all vsRNAs with common 5′ or 3′ ends among the 20- to 29-nt classes, subsequently added their respective flanking genomic sequences (10 nt at each end of the vsRNAs), and performed a logo analysis. We were able to identify the degenerate motif of DVNV̂DC at the cut site (^) of vsRNAs derived both from positive and negative viral strands (Fig. 8A). Enrichments of A in the −3/n−2 position, of G in the −2/n−1 position, and of A in the 2/+2 position are also indicated.
FIG 8.
Enrichment of the motif DVNV̂DC at the boundary of the cleavage (^) of 20- to 29-nt vsRNAs. (A) Logos of 20- to 29-nt vsRNAs with common 5′ (left) or 3′ ends (right) and their flanking sequences. At the cleavage sites of the 5′ end, −1, −2, and −3 denote the first three bases of the viral genome upstream of the 5′ end of the vsRNA (marked with a “1”). At the cleavage sites of the 3′ end, +1, +2, and +3 denote the first three bases of the viral genome downstream of the 3′ end of the vsRNAs (marked with an “n”). Enrichments of base A in the −3/n−2 position, G in the −2/n−1 position, and A in 2/+2 position are also indicated. (B) Statistical analysis of the occurrence of the degenerate motif at the ends of the vsRNA reads in the BmCPV genome (blue bars). A similar analysis of the occurrence of the motif at the ends of vsRNAs shifted by one nucleotide is indicated by the red bars. The vertical axis indicates the value of r, Pearson coefficient of correlation. r values that are higher than 0.073 indicate a positive correlation that is statistically significant (for 22- to 29-nt vsRNAs but not for 20- to 21-nt vsRNAs). The shifted vsRNA ends are negatively correlated.
To verify the enrichment of the degenerate motif for the individual 20- to 29-nt vsRNAs classes, all BmCPV genomic sites matching the degenerate motif were identified and tested for a correlation with the vsRNA reads via Pearson one-tailed tests. As shown in Fig. 8B (blue bars), the vsRNAs sites from all classes except the 20- and 21-nt classes are positively correlated with the existence of the degenerate motif. On the other hand, a single nucleotide shift of one of the two binary matrices under examination (see Materials and Methods) annihilates this effect (Fig. 8B, red bars). Our data therefore suggest that the various vsRNA classes' profiles are related to the cleavage of the degenerate motif by an unknown RNase, which may also be related to the defense against CPV infection.
The failure to identify a correlation between the individual 20- to 21-nt vsRNA classes and the degenerate motif is not unexpected since these classes are further characterized by the abundant Dicer-2-produced vsRNAs, which generate a too-high background. We hypothesize that in the process of viral replication, the host's RNases may recognize and process locally exposed annealed viral sequences at the degenerate motif. Initial cleavage may destabilize transcription/replication and in turn recruit additional RNase activities at these sites that contribute to the genesis of hot spots. Given the similarity of distributions between the 20-nt and the 25- to 29-nt vsRNAs (Fig. 7), we also propose that Dicer-2 could act upon these initially processed templates, extending the processing of dsRNA initiated at the primary vsRNA hot spots.
DISCUSSION
The study of vsRNAs is important to elucidate the interaction between viral pathology and host defenses, particularly for those host-virus systems characterized by an operative exo-RNAi or other pathways that target viral RNA. To our knowledge, this is the first in-depth vsRNA analysis of a Cypovirus infection and of an RNA virus infecting Lepidoptera.
The availability of persistently infected silkworms with BmCPV allowed the comparative study between the persistent and pathogenic infection conditions. Our data indicate that vsRNAs in persistently infected larvae are found in very low abundance, which raises some uncertainty regarding the functionality of these molecules. However, the peak of 20 nt in the vsRNA distribution (22) indicates that the low viral loads during persistent infection are also targeted by Dicer-2 and that the low levels of vsRNAs thus generated represent an adequate defense response. In this defense response, pathogenicity is prevented, although RNAi (and other pathways) are not able to completely clear the virus, which results in the establishment of a persistent infection (22). Persistent infection, however, does not confer substantial protection from subsequent pathogenic infection, at least at the viral dose used in our experiments, since this dose caused considerable developmental and growth defects and may ultimately be lethal. This is in line with (35), where it was also found that previous exposure to an RNA virus does not confer protection against subsequent infection by the same virus in Drosophila. Thus, exo-RNAi is involved in controlling persistent infection by BmCPV, but pathogenicity can be established when a threshold of viral load is reached. This threshold for the capacity of RNAi to control viral infection may differ among silkworm strains with variable BmCPV resistance and may also depend upon environmental conditions.
To date, several studies have demonstrated that in Diptera (Drosophila, Aedes, Culex, and Culicoides) the predominant vsRNA class generated by Dicer-2 is 21 nt in length (4–8, 10–13, 15, 25, 36). In Hemiptera (Laodelphax and Homalodisca) it was shown to be either 21 or 22 nt (9, 14, 37), whereas from a single study it was suggested to be 22 nt in Hymenoptera (Apis) (38). In contrast, we showed that in Bombyx Dicer-2 generates mainly 20-nt vsRNAs. Other observations indicate that the 20-nt class of vsRNAs is lepidopteran-specific with regard to the predominant product of Dicer-2. Jayachandran et al. (16) found abundant 20-nt vsRNAs following infection of Helicoverpa armigera (Lepidoptera; Noctuidae) with the H. armigera single nucleopolyhedrovirus (HaSNPV). In the lepidopteran Sf9 cell line, the predominant size class of baculovirus-derived vsRNAs was also 20 nt in length (39).
Although a limited number of asiRNAs were tested (four in each category of hot, cold, and moderate spots), our experiments to silence the transcript of S10 nevertheless suggest that there is no functional distinction between vsRNAs mapped to hot or cold spots. Excluding the exceptional low activity of m1 and m2 asiRNAs from the analysis, no significant differences were observed among all three classes. Moreover, since moderate spot asiRNAs are expected to have an intermediate effect, the reduced effects of m1 and m2 asiRNAs indicate that less potent 20-nt vsRNAs can be found equally between hot and cold spot-derived vsRNAs. Although a larger number of asiRNAs need to be tested that would preferentially also target transcripts of other segments, our first assessment clearly argues against the different functionality of cold spot and hot spot vsRNAs that were proposed in other infection models (15). When designing the tested asiRNA collection, consideration was given to proposed guidelines for asiRNA design (40) since the base composition near the 5′ and 3′ ends can affect the potency of siRNAs. However, apart from this empirical formula, there is no coherent model theory for predicting siRNA efficacies, and other, as-yet-unidentified elements of the siRNA sequence may be critical.
The distributions of vsRNAs and statistical analysis of the correlation of the mapping of positive-strand and negative-strand 19- to 21-nt siRNAs implicated viral dsRNA as the principal template for Dicer-2-dependent production of the 19- to 21-nt vsRNAs. However, it is not clear at which stage of the infection cycle dsRNA is recognized by Dicer-2. It is known that reoviral genomic dsRNA is tightly packaged and capsid enclosed throughout the cycle of the virus, including during transcription of viral mRNA, which should preclude access by host's Dicer-2 or by other RNases. Thus, genomic dsRNAs in completely or even partially assembled virions are unlikely to be the targets for Dicer-2 processing. Even if such access by RNases is possible, one would expect approximately equal numbers of vsRNAs (normalized by segment's length) derived from the various segments, assuming that all 10 viral segments are represented in each virion. Our data instead indicate that the BmCPV segments are differentially targeted by Dicer-2. For example, S9 is heavily targeted, whereas the larger S7 and S8 are not (Fig. 2 and 3). The very complex process by which reoviral dsRNA replication occurs within the synchronously assembled capsids is far from being completely understood (see reference 41 and references therein), whereas for the genus Cypovirus in particular, studies are scarce (21). Interestingly, it has been indicated that reoviral replication and genome assembly begin with the replication of the smallest of the viral segments and then proceed in a nonrandom fashion, i.e., from the smaller segments to the larger ones (42). Thus, individual replicating BmCPV segments may be differently exposed to Dicer-2 and to other RNases, which could explain why not all segments are targeted at equal frequency.
The existence of segment-specific replication factories in the viral stroma of the infected cells could also provide an explanation for the strong positive-strand bias of the small 16- to 17-nt vsRNA originating from the 3′ ends of segments 4, 7, and 8 (Fig. 2). The 25- to 29-nt vsRNAs derived from half of BmCPV segments also exhibit a positive-strand bias at the 3′ ends of the segments (data not shown). Reoviral dsRNA initiates its replication from a panhandle structure which forms upon 5′-3′ cyclization of the positive strand and is characterized by a single-stranded 3′ tail (43). The vsRNAs that map to the 3′ of the positive strands may be derived from regulatory (panhandle) sequences during BmCPV dsRNA segment replication.
Of particular interest is the finding of the degenerate motif DVNV̂DC at the cut site of vsRNAs suggesting either the function of a yet unidentified RNase or a new function/preference of a known RNase. Because distributions of 19- to 21-nt vsRNAs produced by Dicer-2 are similar to those of larger vsRNAs (25 to 29 nt), it was hypothesized that this RNase activity is exerted on viral replication intermediates, as in the case of Dicer-2 (see above). Our analysis suggests that the identified motif correlates with the vsRNA distributions and therefore that the corresponding RNase activity may be an important early factor for vsRNA biogenesis and distribution. We propose that initial endoribonucleolytic cleavages by other enzymes may facilitate Dicer-2 in gaining access to the numerous hot spots, although we cannot rule out the possibility that Dicer-2 may itself be implicated in the initial endo-RNase activities. Interestingly, mammalian RNase III class enzymes Dicer and Drosha, both implicated in the microRNA pathway, have been shown to exhibit specific base preferences at the cut site (44), which nevertheless do not match our motif. Further, no eukaryotic RNases with sequence specificity have been identified to date (45), whereas the unique case of RNase Mini-III from Bacillus subtilis that cleaves dsRNA in a sequence-specific manner was discovered recently (46). Clearly, more studies are required to elucidate the mechanism and to identify the responsible endonuclease(s), which also warrants research in other insects and organisms.
The positive strand of S10 stands out for the very high RPKM values (Fig. 1) and for the concomitant strong bias of the corresponding (+)-vsRNAs (Fig. 2), clearly suggesting that the high expression of polyhedrin mRNA creates an additional source for vsRNAs. A recent study indicates that such bias can be created by sequestration of (−)-vsRNAs by abundant viral positive strands (47). However, the profile of S10 (+)-vsRNAs is very distinct because of the existence of the very abundant 25- to 30-nt classes (Fig. 2) that are not reflected in the (−)-vsRNAs profile (Fig. 2). Furthermore, sequestration of (−)-vsRNAs to polyhedrin transcripts should result in cleavage or inhibition of translation, while instead polyhedra accumulate in the infected larvae (22). Excluding this possibility, the question arises as to which mechanism is responsible for the production of the abundant (+)-vsRNAs of S10. As shown in Fig. 2, the distribution profile of vsRNAs from the positive strand of S10 has two peaks of abundance: one peak of around 20 nt and a second one of around 28 nt (the 2inf sample). For the peak around 20 nt, we propose that this reflects processing by Dicer-2 since this enzyme can also process structured ssRNA in both Drosophila and mosquitoes (4). Compared to the vsRNAs produced from viral dsRNA, the less sharp decrease in reads of vsRNA classes flanking the 20-nt class may indicate the mismatches and small bulges commonly found in structured RNA that should render Dicer-2 products less of a definite length.
Another and not mutually exclusive possibility is that intramolecular stem-loops of viral mRNA are targeted via a noncanonical miRNA pathway. Such a pathway could presumably involve Argonaute-2, Argonaute-1, and Dicer-1, since it has been demonstrated in mosquito cells infected with the Dengue virus 2 (48). In that study, the authors provide evidence that a miRNA-like molecule derived from the RNA virus autoregulates viral replication. Interestingly, we also found a pre-miRNA-like stem-loop from the positive strand of segment 10 that modestly downregulates MycHis-tagged polyhedrin protein in cotransfection experiments. Clearly, more studies are required for deciphering whether this and/or other BmCPV intramolecular hairpins are produced via an atypical miRNA pathway and whether they play an important role in viral autoregulation.
The observed peak of 28 nt for the (+)-vsRNAs derived from segment 10 (Fig. 2) cannot be convincingly explained either by Dicer-2 processing or by assuming an involvement of miRNA-related pathways since the miRNAs (20 to 27 nt in size) have the highest frequencies: approximately 20 to 23 nt (49). A distinct class of larger vsRNAs (∼27 nt) that are implicated in Dicer-independent antiviral RNAi in Diptera is the class of the so-called Piwi-interacting small RNAs (piRNAs) (8, 50, 51). However, piRNAs typically bear specific signatures such as a U at position 1 or as an A at position 10 of their sequences (reflecting the ping-pong signature [8, 50, 51]), but piRNA signatures are absent from our deep-sequencing data of BmCPV-derived vsRNA. We thus hypothesize that the abundant vsRNAs that are approximately 26 to 30 nt in length, which are derived from positive-strand of segment 10, are produced by a yet-to-be-defined mechanism.
ACKNOWLEDGMENTS
This study was performed in the framework of the Target Identification for Disease Diagnosis and Treatment (DIAS) project within GSRT's KRIPIS action, funded by Greece and the European Regional Development Fund of the European Union under the O.P. Competitiveness and Entrepreneurship (NSRF 2007-2013). G.S. was also supported by the Fund for Scientific Research (FWO-Vlaanderen, Belgium).
We thank H. Mori (Kyoto Institute of Technology, Japan), who generously provided the purified BmCPV polyhedra used for the infections.
REFERENCES
- 1.Bronkhorst AW, van Rij RP. 2014. The long and short of antiviral defense: small RNA-based immunity in insects. Curr Opin Virol 7:19–28. doi: 10.1016/j.coviro.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 2.van Rij RP, Berezikov E. 2009. Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol 17:163–171. doi: 10.1016/j.tim.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Jinek M, Doudna JA. 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 4.Sabin LR, Zheng Q, Thekkat P, Yang J, Hannon GJ, Gregory BD, Tudor M, Cherry S. 2013. Dicer-2 processes diverse viral RNA species. PLoS One 8:e55458. doi: 10.1371/journal.pone.0055458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. 2010. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis 4:e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynt A, Liu N, Martin R, Lai EC. 2009. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci U S A 106:5270–5275. doi: 10.1073/pnas.0813412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leger P, Lara E, Jagla B, Sismeiro O, Mansuroglu Z, Coppee JY, Bonnefoy E, Bouloy M. 2013. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J Virol 87:1631–1648. doi: 10.1128/JVI.02795-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Andika IB, Shen J, Lv Y, Ji Y, Sun L, Chen J. 2013. Characterization of rice black-streaked dwarf virus- and rice stripe virus-derived siRNAs in singly and doubly infected insect vector Laodelphax striatellus. PLoS One 8:e66007. doi: 10.1371/journal.pone.0066007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. 2008. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci U S A 105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnettler E, Ratinier M, Watson M, Shaw AE, McFarlane M, Varela M, Elliott RM, Palmarini M, Kohl A. 2013. RNA interference targets arbovirus replication in Culicoides cells. J Virol 87:2441–2454. doi: 10.1128/JVI.02848-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, Ebel GD, Olson KE, Blair CD. 2010. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl Trop Dis 4:e848. doi: 10.1371/journal.pntd.0000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Cleef KW, van Mierlo JT, Miesen P, Overheul GJ, Fros JJ, Schuster S, Marklewitz M, Pijlman GP, Junglen S, van Rij RP. 2014. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res 42:8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nandety RS, Fofanov VY, Koshinsky H, Stenger DC, Falk BW. 2013. Small RNA populations for two unrelated viruses exhibit different biases in strand polarity and proximity to terminal sequences in the insect host Homalodisca vitripennis. Virology 442:12–19. doi: 10.1016/j.virol.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Siu RW, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, Barry G, Attarzadeh-Yazdi G, Rodriguez-Andres J, Nash AA, Merits A, Fazakerley JK, Kohl A. 2011. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J Virol 85:2907–2917. doi: 10.1128/JVI.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayachandran B, Hussain M, Asgari S. 2012. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J Virol 86:13729–13734. doi: 10.1128/JVI.02041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arella M, Lavallee C, Belloncik S, Furuichi Y. 1988. Molecular cloning and characterization of cytoplasmic polyhedrosis virus polyhedrin and a viable deletion mutant gene. J Virol 62:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Jin L, Zhou ZH. 2008. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagiwara K, Rao S, Scott SW, Carner GR. 2002. Nucleotide sequences of segments 1, 3 and 4 of the genome of Bombyx mori cypovirus 1 encoding putative capsid proteins VP1, VP3, and VP4, respectively. J Gen Virol 83:1477–1482. doi: 10.1099/0022-1317-83-6-1477. [DOI] [PubMed] [Google Scholar]
- 20.Mori H, Metcalf P. 2010. Cypoviruses, p 307–323. In Asgari S, Johnson K (ed), Insect virology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 21.Yang J, Cheng Z, Zhang S, Xiong W, Xia H, Qiu Y, Wang Z, Wu F, Qin CF, Yin L, Hu Y, Zhou X. 2014. A cypovirus VP5 displays the RNA chaperone-like activity that destabilizes RNA helices and accelerates strand annealing. Nucleic Acids Res 42:2538–2554. doi: 10.1093/nar/gkt1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolliopoulou A, Van Nieuwerburgh F, Stravopodis DJ, Deforce D, Swevers L, Smagghe G. 2015. Transcriptome analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PLoS One 10:e0121447. doi: 10.1371/journal.pone.0121447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J Comput Graph Statist 5:299–314. doi: 10.1080/10618600.1996.10474713. [DOI] [Google Scholar]
- 24.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marques JT, Wang JP, Wang X, de Oliveira KP, Gao C, Aguiar ER, Jafari N, Carthew RW. 2013. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog 9:e1003579. doi: 10.1371/journal.ppat.1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douris V, Swevers L, Labropoulou V, Andronopoulou E, Georgoussi Z, Iatrou K. 2006. Stably transformed insect cell lines: tools for expression of secreted and membrane-anchored proteins and high-throughput screening platforms for drug and insecticide discovery. Adv Virus Res 68:113–156. doi: 10.1016/S0065-3527(06)68004-4. [DOI] [PubMed] [Google Scholar]
- 27.Li TC, Scotti PD, Miyamura T, Takeda N. 2007. Latent infection of a new alphanodavirus in an insect cell line. J Virol 81:10890–10896. doi: 10.1128/JVI.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA package 2.0. Algorithms Mol Biol 6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z. 2007. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res 35:W339–W344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson R, Meidinger RG, Iatrou K. 1992. A cellular promoter-based expression cassette for generating recombinant baculoviruses directing rapid expression of passenger genes in infected insects. Virology 190:815–823. doi: 10.1016/0042-6822(92)90919-G. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Swevers L, Iatrou K, Huvenne H, Smagghe G. 2012. Bombyx mori DNA/RNA non-specific nuclease: expression of isoforms in insect culture cells, subcellular localization and functional assays. J Insect Physiol 58:1166–1176. doi: 10.1016/j.jinsphys.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Georgomanolis T, Iatrou K, Swevers L. 2009. BmCAP, a silkmoth gene encoding multiple protein isoforms characterized by SoHo and SH3 domains: expression analysis during ovarian follicular development. Insect Biochem Mol Biol 39:892–902. doi: 10.1016/j.ibmb.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro JS, Langlois RA, Pham AM, Tenoever BR. 2012. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA 18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longdon B, Cao C, Martinez J, Jiggins FM. 2013. Previous exposure to an RNA virus does not protect against subsequent infection in Drosophila melanogaster. PLoS One 8:e73833. doi: 10.1371/journal.pone.0073833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, Perot J, Pfeffer S, Hoffmann JA, Saleh MC, Imler JL. 2010. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci U S A 107:19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Huang L, Wang Z, Fu S, Che J, Qian Y, Zhou X. 2014. Identification of Himetobi P virus in the small brown planthopper by deep sequencing and assembly of virus-derived small interfering RNAs. Virus Res 179:235–240. doi: 10.1016/j.virusres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Chejanovsky N, Ophir R, Schwager MS, Slabezki Y, Grossman S, Cox-Foster D. 2014. Characterization of viral siRNA populations in honey bee colony collapse disorder. Virology 454-455:176–183. doi: 10.1016/j.virol.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Mehrabadi M, Hussain M, Matindoost L, Asgari S. 2015. The baculovirus antiapoptotic p35 protein functions as an inhibitor of the host RNA interference antiviral response. J Virol 89:8182–8192. doi: 10.1128/JVI.00802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi J, Mizoguchi T, Fujiwara H. 2011. siRNAs induce efficient RNAi response in Bombyx mori embryos. PLoS One 6:e25469. doi: 10.1371/journal.pone.0025469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boudreaux CE, Kelly DF, McDonald SM. 2015. Electron microscopic analysis of rotavirus assembly-replication intermediates. Virology 477:32–41. doi: 10.1016/j.virol.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung PY, Roy P. 2014. Sequential packaging of RNA genomic segments during the assembly of Bluetongue virus. Nucleic Acids Res 42:13824–13838. doi: 10.1093/nar/gku1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, Patton JT. 1998. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J Virol 72:7387–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starega-Roslan J, Witkos TM, Galka-Marciniak P, Krzyzosiak WJ. 2015. Sequence features of Drosha and Dicer cleavage sites affect the complexity of isomiRs. Int J Mol Sci 16:8110–8127. doi: 10.3390/ijms16048110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomecki R, Dziembowski A. 2010. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA 16:1692–1724. doi: 10.1261/rna.2237610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glow D, Pianka D, Sulej AA, Kozlowski LP, Czarnecka J, Chojnowski G, Skowronek KJ, Bujnicki JM. 2015. Sequence-specific cleavage of dsRNA by Mini-III RNase. Nucleic Acids Res 43:2864–2873. doi: 10.1093/nar/gkv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris CJ, Molnar A, Muller SY, Baulcombe DC. 2015. FDF-PAGE: a powerful technique revealing previously undetected small RNAs sequestered by complementary transcripts. Nucleic Acids Res 43:7590–7599. doi: 10.1093/nar/gkv604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussain M, Asgari S. 2014. MicroRNA-like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc Natl Acad Sci U S A 111:2746–2751. doi: 10.1073/pnas.1320123111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Cai Y, Ye L, Wang S, Che J, You Z, Yu J, Zhong B. 2014. MicroRNA expression profiling of the fifth-instar posterior silk gland of Bombyx mori. BMC Genomics 15:410. doi: 10.1186/1471-2164-15-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. 2012. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog 8:e1002470. doi: 10.1371/journal.ppat.1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Q, Luo Y, Lu R, Lau N, Lai EC, Li WX, Ding SW. 2010. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci U S A 107:1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]