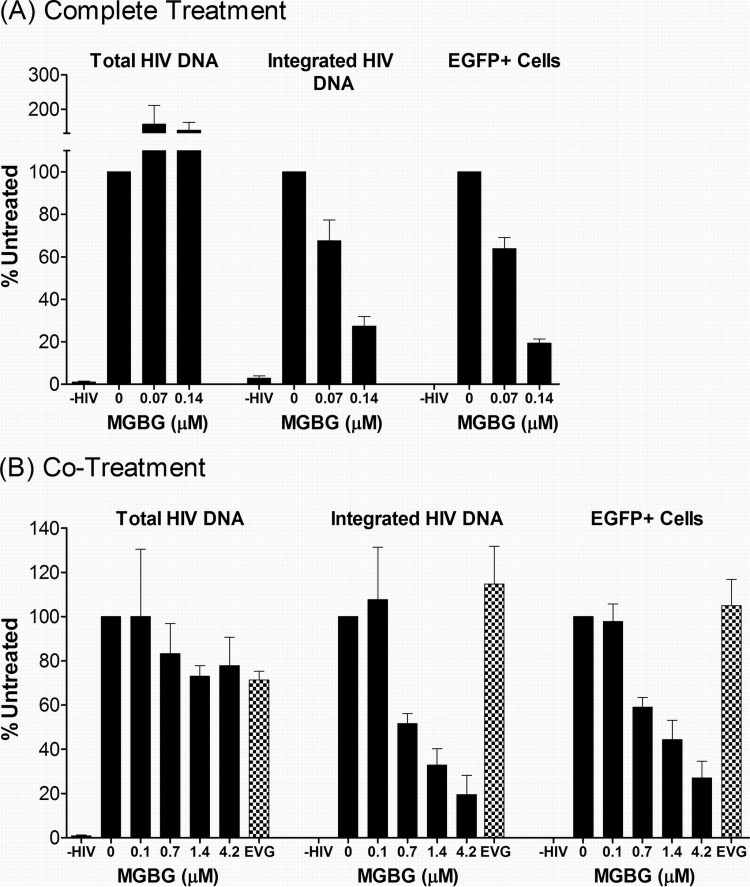
FIG 6.
Inhibition of HIV DNA integration in MGBG-treated cells. Human macrophages were infected with EGFP-tagged HIV and treated with MGBG in either the “complete treatment” mode or “cotreatment” mode as indicated. At the end of the experiments, a fraction of the cells were used for FACS analysis of EGFP expression, while the rest were used for DNA extraction. Real-time PCR was used to quantify HIV DNA in the total or high-molecular-weight DNA. The copy numbers of β-globin in the samples were simultaneously determined to calculate the load or relative burden of the HIV DNA. Noninfected cells were included as a control for the specificity of the assays. The data for the complete treatment and cotreatment modes were from 7 and 3 donors, respectively. Due to individual variations, all data were normalized against an HIV-infected, nontreated sample in each experiment. The graphs show the means and SEMs of the normalized results from multiple donors. The P values for a linear trend in the association between MGBG dose and effect on proviral DNA were 0.045 and 0.073 for the complete and cotreatment modes, respectively. Detailed FACS data for EGFP expression are shown in Fig. S4 and S5 in the supplemental material.