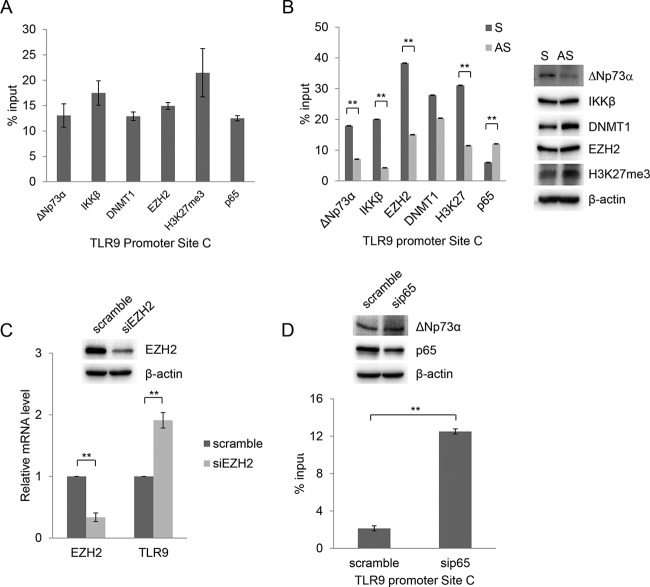
FIG 4.
ΔNp73α and p65 compete for binding to the TLR9 promoter. (A) HPV38 E6/E7 HFK were processed for ChIP using ΔNp73α, IKKβ, DNMT1, EZH2, H3K27me3, and p65 antibodies. qPCR was performed using specific primers flanking NF-κB RE site C within the TLR9 promoter. Data are the means from three independent experiments. All the bindings shown are statistically significant compared to the negative control, with a P value of <0.01. (B) HPV38 E6/E7 HFK were transfected with ΔNp73α S and AS oligonucleotides, and the levels of indicated cellular proteins were determined by IB (right side). Total cellular extracts were processed for ChIP with the indicated antibodies. Data are from one representative experiment of two independent experiments. **, P < 0.01. (C) HPV38 E6/E7 HFK were transfected with a control siRNA (scramble) or siRNA specific for EZH2 (siEZH2). After 72 h, total RNA was extracted and EZH2 or TLR9 mRNA levels were analyzed by RT-qPCR. Error bars represent standard deviations from three biological replicates. **, P < 0.01. The inset shows the EZH2 protein levels determined by IB. (D) HPV38 E6/E7 HFK were transfected with a control siRNA (scramble) or siRNA specific for p65 (sip65), and p65 as well as ΔNp73a levels were determined by IB (inset). Total cellular extracts were processed for ChIP using anti-ΔNp73α antibody. The results were analyzed by qPCR. Data are the means from three independent experiments. **, P < 0.01.