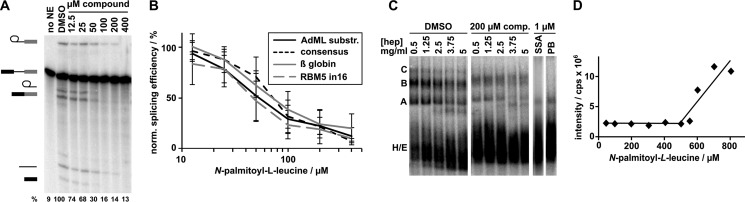
FIGURE 4.
N-palmitoyl-l-leucine inhibits splicing of a variety of substrates and interferes with late spliceosome assembly. A, representative denaturing gel analysis of RNA isolated from DMSO-treated control splicing reactions or reactions containing indicated concentrations of N-palmitoyl-l-leucine. Identities of the bands are schematized as (from top to bottom) lariat-intermediate, pre-mRNA, free lariat, mRNA, free intron, 5′ exon intermediate. Normalized splicing efficiency is shown at the bottom of each lane. B, average normalized splicing efficiency plotted versus N-palmitoyl-l-leucine concentration for four pre-mRNA substrates with different branch region sequences from triplicate splicing reactions. C, native gel analysis of ”kinetically trapped“ spliceosomes assembled in DMSO (first lane of left gel) and spliceosomes assembled in the presence of 200 μm N-palmitoyl-l-leucine (first lane of middle gel) or 1 μm SSA and pladienolide B (PB), which are splicing inhibitors that arrest at early assembly stages (right gels). The 2nd-5th lanes of the first gels show complexes challenged with increasing concentrations of heparin (hep) at the indicated concentration. D, dynamic light scattering analysis of N-palmitoyl-l-leucine plotting scattering intensity versus increasing compound concentration.