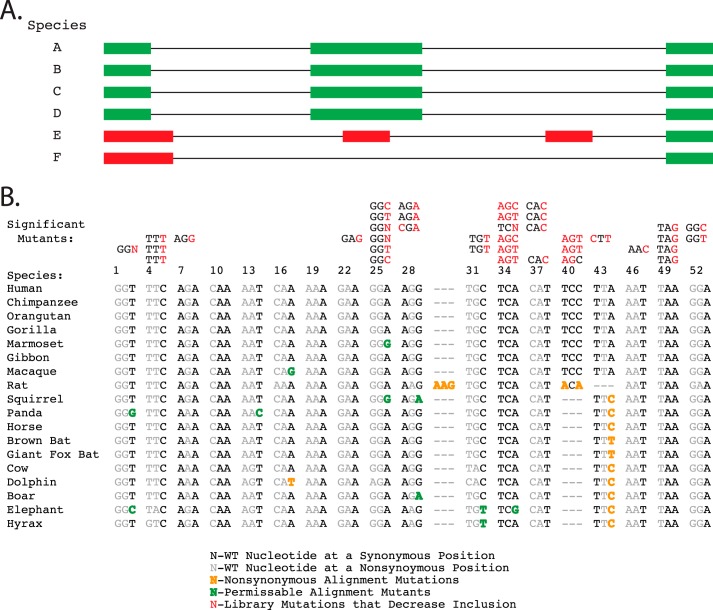
FIGURE 4.
Sequence alignment of SMN1 exon 7 with 17 animals with similar intron/exon architecture. A, a graphical representation of the method used to filter species alignments based on conservation of intron/exon architecture. Species E and F would not be included in an architecture-filtered alignment. B, the alignment of similar mammals that pass the intron/exon architecture filter to human SMN1 exon 7. The mutations that caused significant decreases in the fitness index value are listed above their position in the alignment with the specific hexamer mutations in red. The alignment below the hexamers shows the silent mutation positions that diverge from the human sequence. The mutations that did not cause changes in the fitness index value are highlighted in green. The color-coding used to differentiate the effects of the observed mutants is shown in the key on the figure bottom. The category “nonsynonymous alignment mutations” refers to positions in the multispecies alignment where observed nucleotide substitutions would be considered nonsynonymous mutations in the human exon.