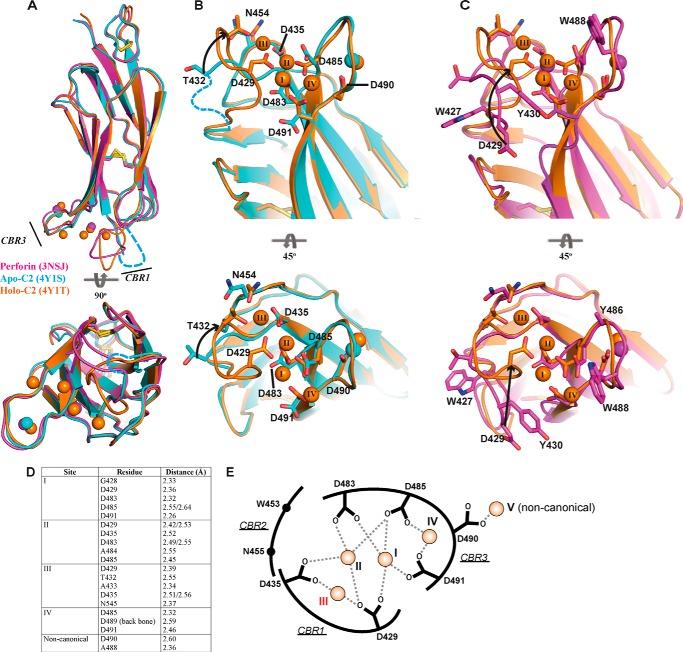
FIGURE 2.
Crystal structure of apo- and holo-C2 quad(410–535) superimposed with the C2 domain from full-length perforin. A, C2 domain of 3NSJ (magenta) superimposed with both the apo-C2 (cyan) and holo-C2 (orange) quad(410–535). CBRs 1 and 3 are identified, and the corresponding colored spheres represent the Ca2+ ions for each structure. The view is rotated by 90° to view the C2 domain from the membrane perspective. CBR1 in the apo-C2 quad(410–535) structure is disordered, and the loop was not built into the density (cyan dashed line). B, superimposed structures of the apo-C2 (cyan) and holo-C2 (orange) quad(410–535) to illustrate the re-organization that occurs in the residues involved in Ca2+ coordination. The residues from Ala-427 to Ala-431 (CBR1, cyan dashed line) were not visible in the electron density map of the apo-structure and were not modeled into the final structure. Significant movement is observed in the CBR1, where residue Thr-432 moves through 6.8 Å, after which the CBR1 loop becomes well ordered and visible in the holo-C2 structure. Upon movement of the loop, Asp-429 is re-positioned to engage Ca2+ in positions I–III. C, superimposition of the C2 domain from murine perforin and the holo-C2 quad(410–535) further demonstrates the significant movement of residue Asp-429 over 11 Å. Throughout, key residues are represented in stick form labeled by residue number; arrows indicate movement, and disulfide bonds are represented as yellow sticks. Ca2+ is numbered as described previously (17), and the additional observed Ca2+ is numbered sequentially (position IV). D, key residues of the holo-C2 quad(410–535) structure making contact with Ca2+. E, schematic representation of Ca2+-binding interactions in the CBRs. The coordinates of the side chain carboxyl groups of conserved Asp residues and Asp-490 with Ca2+ ion at each site are indicated as dashed lines. The Ca2+ ions at each site are shown as orange spheres with corresponding site numbers. The weakest affinity site (III) is labeled in red.