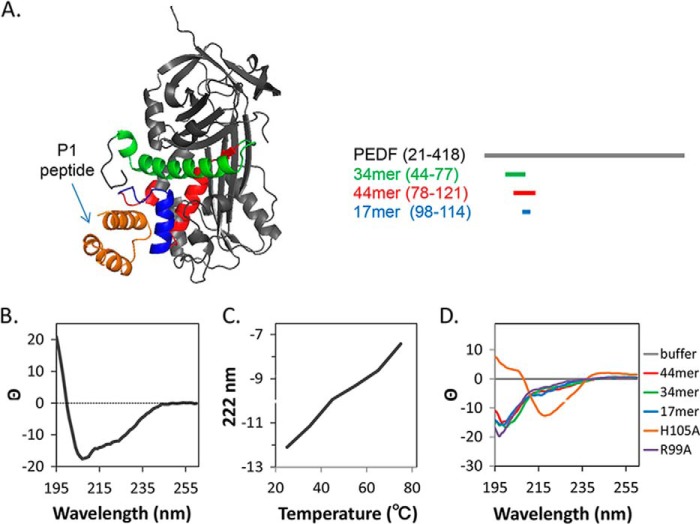
FIGURE 1.
Modeling of binding interaction of LBD of PEDF-R to PEDF. A, the structure of the LBD peptide P1 derived from the ab initio fragment assembly protocol of Rosetta is shown in orange. The resultant P1 peptide structure docked to the PEDF crystal structure (Protein Data Bank code 1IMV) using Rosetta program is shown. P1 peptide docked to a cleft that contained a solvent-exposed region corresponding to α-helix C within the residues 98–114 (17-mer; blue) of the neurotrophic 44 amino acid region (red). The antiangiogenic peptide region (34-mer; green) was not part of the docking region. A linear representation of the PEDF structure is given at right with identical color code as above. B, the CD spectrum of peptide P1 contained the negative ellipticity at 222 and 208 nm and a positive peak at 190 nm that are indicative of α helical structure. C, additionally, the CD spectrum of peptide P1 changed to reflect peptide unfolding as the temperature was increased from 25 to 80 °C, indicating that the P1 fragment is folded at 25 °C and unfolds at higher temperatures. D, CD spectra of peptides used in this study are shown.