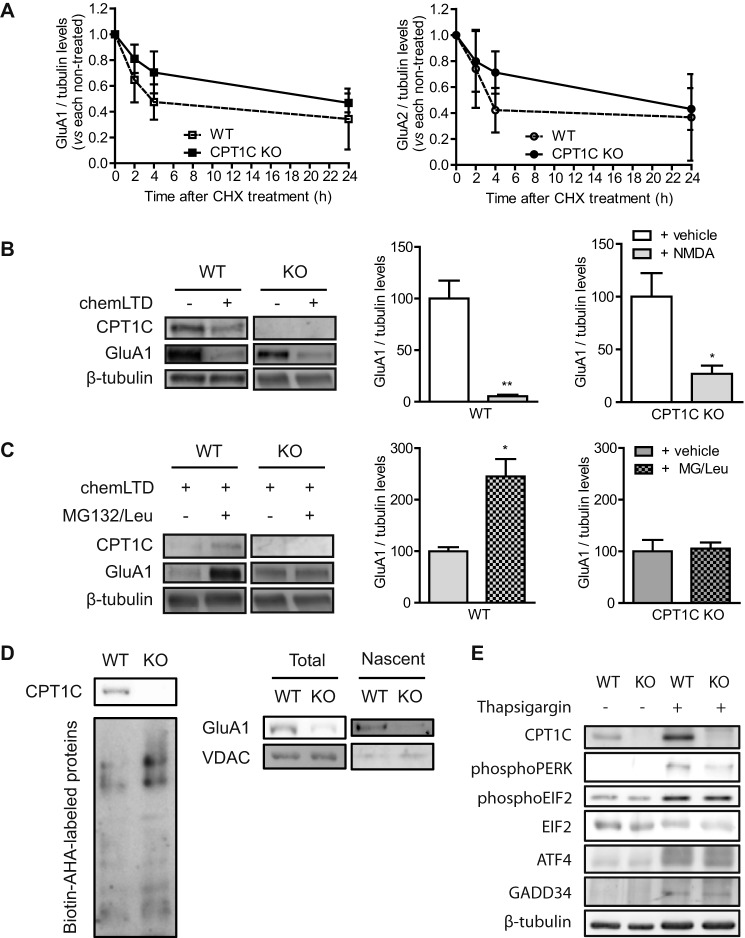
FIGURE 7.
CPT1C effects on AMPAR protein degradation and synthesis. A, basal degradation of AMPARs. At 14 DIV, neurons were treated with cycloheximide (10 μm) for 24 h to inhibit protein translation. Protein levels were analyzed by immunoblot, and results were normalized to WT and CPT1C KO non-treated cells. Values are shown as the mean ± S.E. (error bars) in four independent experiments. B, NMDA-dependent degradation of GluA1. 14 DIV-cultured neurons were stimulated with NMDA (50 μm) for 5 min (chemLTD), and the medium was changed. Five hours later, cells were lysated, and protein levels were analyzed. C, GluA1 protein synthesis after chemLTD. Five hours after NMDA stimulation, neurons were treated with MG132 (10 μm) and leupeptin (200 μg/ml) for an additional 3 h. GluA1 accumulation was evaluated. The graphs show the values as mean ± S.E. in three independent experiments. D, de novo synthesis of GluA1. Total lysates (left) were obtained to analyze total biotin-azidohomoalanine-labeled proteins. GluA1 and voltage-dependent anion-selective channel (VDAC) (as control) were analyzed by immunoblotting in cell extracts (total) and in NeutrAvidin-bound fractions (nascent proteins; right). The image is representative of two independent experiments. E, analysis of endoplasmic reticulum stress markers in WT or CPT1C KO neuronal cultures. CPT1C, phospho-PERK, phospho-EIF2, ATF4, and GADD34 protein levels were determined by immunoblotting. Thapsigargin (ER stress inductor; 5 μm for 24 h) was used as a positive control. *, p < 0.05; **, p < 0.01.