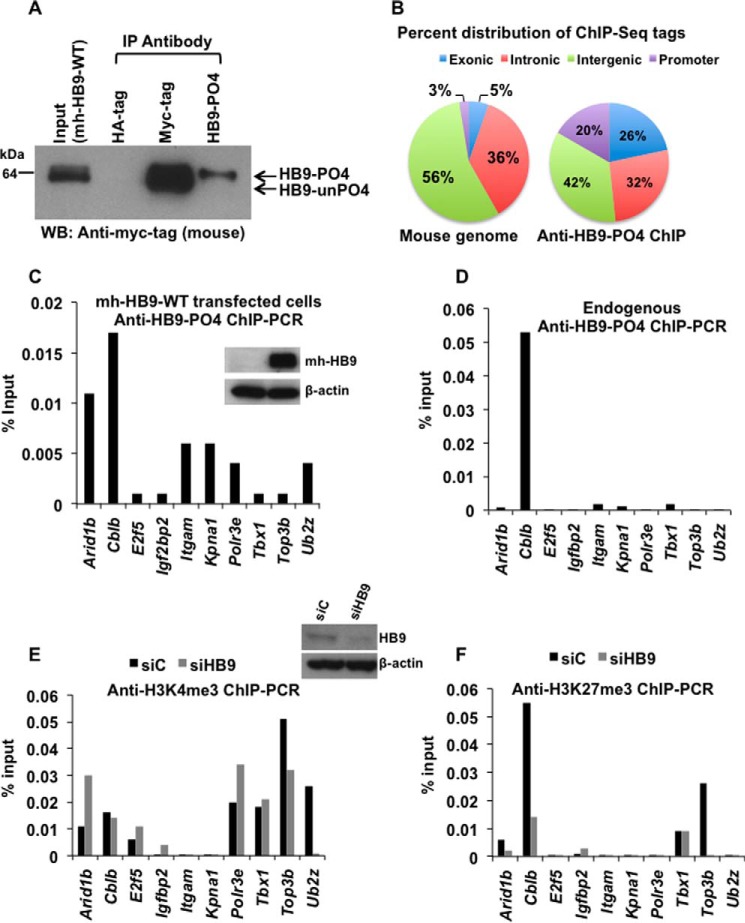
FIGURE 3.
Identification of Cblb as a phospho-HLXB9 target gene. A, the anti-HB9-PO4 antibody specifically recognizes the phosphorylated isoform of HLXB9. WCE and chromatin were prepared from MIN6-4N cells transfected with a plasmid expressing myc-his-tagged HLXB9 (mh-HB9-WT). WCE was used IP with rabbit antibodies anti-myc-tag or anti-HB9-PO4 and detected by Western blot (WB) with mouse anti-myc-tag. Rabbit anti-HA-tag was used as the negative control. The input WCE and anti-myc-tag IP display a doublet corresponding to phospho-HLXB9 and unphosphorylated HLXB9. Anti-HB9-PO4 could specifically immunoprecipitate phospho-HLXB9 corresponding to the top band of the doublet. B, significant enrichment of promoter regions among the anti-HB9-PO4 ChIP-Seq tags. Chromatin prepared from MIN6-4N cells transfected in A was used for ChIP with anti-HB9-PO4. DNA obtained before and after ChIP was used for preparing libraries followed by deep sequencing (ChIP-Seq) and mapping of the anti-HB9-PO4-specific ChIP-Seq tags to the mouse genome. The pie chart shows the percent distribution of tags in the mouse genome (a typical input library, Genomatix) and in the anti-HB9-PO4 ChIP-Seq at the indicated regions; 20% of the anti-HB9-PO4 ChIP-Seq tags were located near promoter regions and selected for further analysis. C, phospho-HLXB9 occupancy is highest at the Arid1b and Cblb gene in cells overexpressing HLXB9. ChIP-quantitative PCR assay for validating the 10 phospho-HLXB9 targets is shown as the percent of input chromatin DNA PCR for each primer pair. Chromatin prepared from MIN6-4N cells expressing mh-HB9-WT was used for anti-HB9-PO4 ChIP. Also shown is a Western blot confirming overexpression of HLXB9 (myc-tag antibody) and β-actin as the loading control. D, endogenous phospho-HLXB9 occupancy is highest at the Cblb gene. ChIP-quantitative PCR assay of the 10 phospho-HLXB9 targets is shown as percent of input chromatin DNA PCR for each primer pair. Chromatin prepared from MIN6-4N cells was used for endogenous anti-HB9-PO4 ChIP. Endogenous phospho-HLXB9 occupancy was only detected at Cblb. E and F, H3K4me3 at Cblb unaffected but reduced H3K27me3 upon HLXB9 knockdown. ChIP-quantitative PCR assay of the 10 phospho-HLXB9 targets is shown as the percent of input chromatin DNA PCR for each primer pair. Chromatin prepared from MIN6-4N cells transfected with control siRNA (siC) or HLXB9 siRNA (siHB9) was used for endogenous anti-H3K4me3 ChIP (E) or H3K27me3 ChIP (F). Also shown is a Western blot confirming knockdown of HLXB9 (HLXB9 antibody) and β-actin as the loading control. In siC versus siHB9, reciprocal H3K4me3 or H3K27me3 at only Cblb was HLXB9 binding-dependent because endogenous phospho-HLXB9 was only found to occupy Cblb (D).