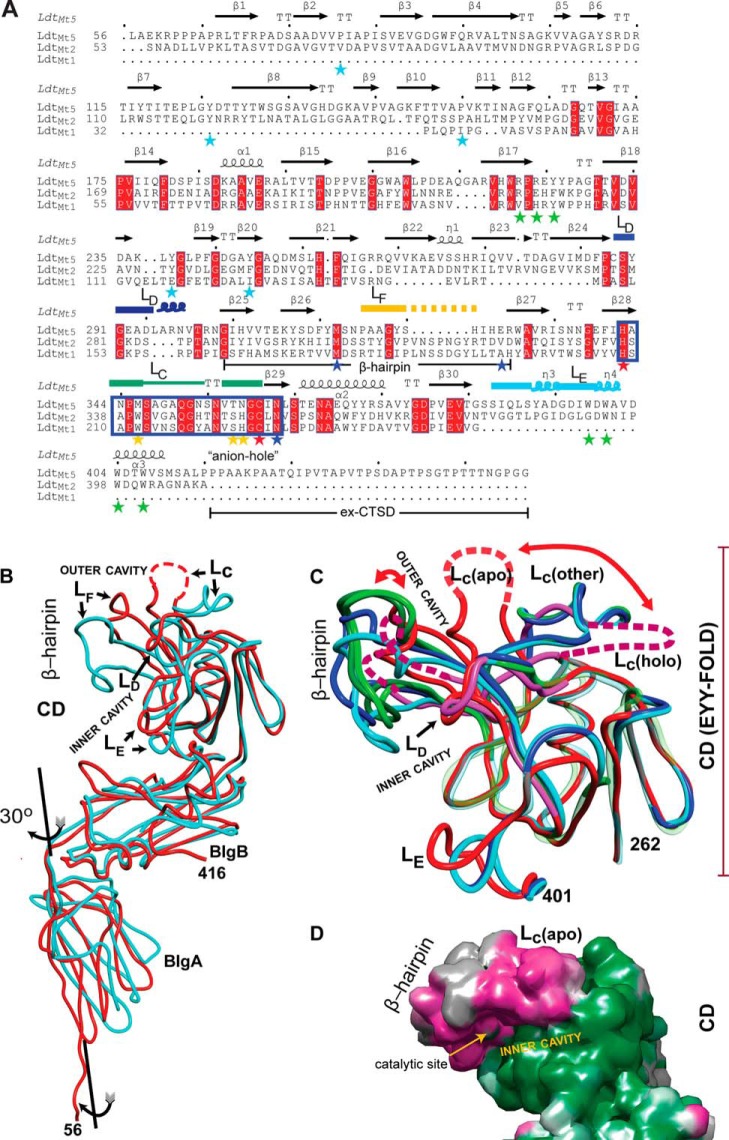
FIGURE 2.
Comparison of M. tuberculosisld-transpeptidases. A, the sequence alignment based on the structural superposition. The observed secondary structures are noted above the amino acid sequences. Starred residues are those highlighted in the text (blue, coordinating His342; cyan, BIgA and BIgB interface; green, CTSD-BIgB-CD core interaction; red, catalytic residues; yellow, loop LD). Named loops are also marked, and the characteristic ld-transpeptidase motif is boxed in blue rectangle. B, overlay of the apo structures of LdtMt2 (Protein Data Bank code 3VYN; in cyan) and LdtMt5 (this work; in red). Observed differences in the CD are concentrated in the β-hairpin and loops LC–LE. Disordered regions are represented as dashed lines. C, the LdtMt5 adduct structure displays the largest CD conformational changes among characterized ld-transpeptidases. The EYY-folded CD cores of apo-LdtMt5 (red), holo-LdtMt5 (pink), and the apo and holo crystal structures of LdtMt1 (Protein Data Bank code 4JMN, apo, in light green; Protein Data Bank code 4JMX, imipenem adduct, in dark green), and LdtMt2 (Protein Data Bank code 3VYN, apo, in cyan; Protein Data Bank code 3VYP, meropenem adduct, in blue) are shown as smoothed Cα traces. Structural elements with high r.m.s.d. are shown in full opacity; the remaining structural elements are displayed as transparent Cα traces to demonstrate conservation of the EYY-folded CD cores. The largest changes among structurally characterized ld-transpeptidases are observed in the β-hairpin, and in the case of LdtMt5, there is a dramatic displacement of loop LC that occurs after adduct formation (indicated with red curved arrows). The C-terminal portion of the CTSDs was excluded for clarity. D, accessible surface map of apo-LdtMt5 colored by the magnitude of the observed atomic temperature factors from low (green) to high (magenta) motility. The flexibility of the β-hairpin as indicated by the high atomic temperature factor correlates with its large displacement upon adduct formation. These images, the sequence alignment, and structural superpositions were performed using the program MOE. The sequence representation was performed using ESPript3 (48).