FIGURE 4.
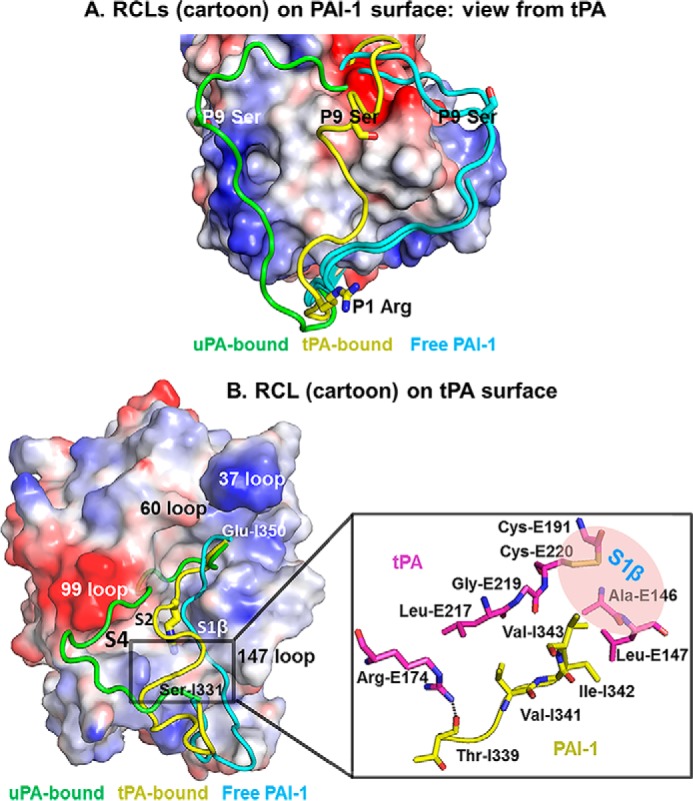
Conformation of the RCL in the tPA·PAI-1 is different from in free PAI-1 and in uPA·PAI-1 Michaelis complexes. A, alignment of active PAI-1 RCL in free form and in PA-bound form. The charged surface representation is the PAI-1 with its RCL removed from the PDB. The RCLs (cyan) of active free PAI-1 (PDB code 1B3K and 1DVM) (26, 33) were observed to adopt an extended conformation just above β-sheet C regardless of the crystal packing and crystallization conditions. The RCL (green) of uPA-bound PAI-1 was observed to flip to another side of PA (PDB code 3PB1) (15). The RCL (yellow) of tPA-bound PAI-1 is at a location between free PAI-1 and uPA-bound. B, left panel, the relative conformations of the RCL in the tPA-SPD·PAI-1 and the uPA-SPD·PAI-1 (PDB-ID 3PB1) Michaelis complexes are displayed on a charged surface representation of tPA-SPD. The RCL in the current structure (yellow) has a localization between the RCL of free PAI-1 (cyan, PDB code 1B3K (33)) and the RCL in the uPA-bound form (green). Right panel, the detailed interactions leading to the kinked conformation of PAI-1. Arg-E174 of tPA (magenta) and Thr-I339 of PAI-1 (yellow) forms a hydrogen bond, which fixes one side of the RCL. The PAI-1 segment Val-341–Ile-342–Val-343 interacts with tPA through hydrophobic interaction, fixing the other end of PAI-1 RCL.
