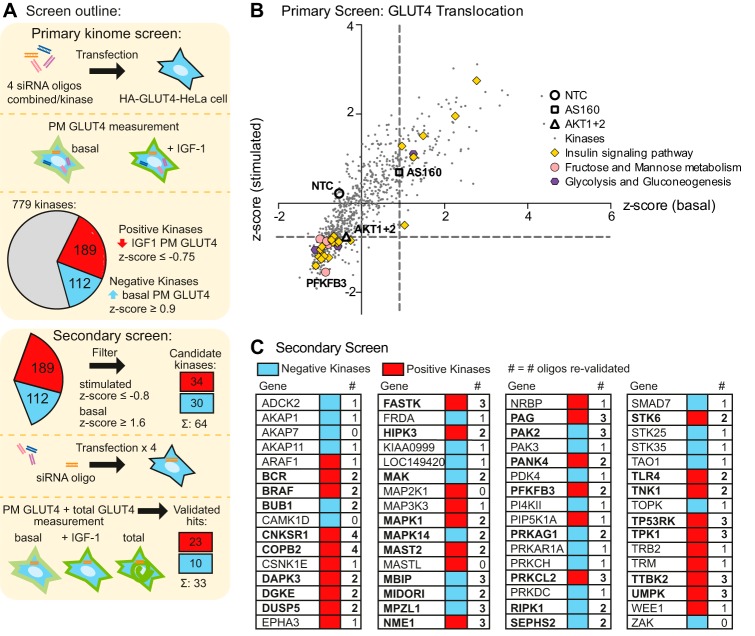
FIGURE 2.
High throughput siRNA screen identifies kinases involved in GLUT4 translocation. A, the screen workflow is shown. In the primary screen, HA-GLUT4-HeLa cells were transfected with siRNA pools targeting 779 kinases and HA-GLUT4 translocation to the PM in response to IGF-1 stimulation was determined as described under “Experimental Procedures.” B, results of the primary screen with siRNA pools targeting 779 kinases are shown. Z-score of stimulated PM GLUT4 was plotted against the Z-score of basal PM GLUT4 for each of the kinases, with 3 controls (NTC, AS160, and Akt1 + 2) indicated. Dashed lines indicate Z-score cut-off to identify hits (Z-score stimulated ≤−0.75 = positive kinases, Z-score basal ≥ 0.9 = negative kinases). Components of enriched KEGG pathways are indicated. C, secondary screen with 4 single siRNAs was performed on 64 candidate kinases (34 positive and 30 negative kinases) and the number of oligos that reproduced the result of the primary screen (greater effect or within 10% of primary screen result) are indicated. Validated kinases (≥2 oligos) are shown in bold.