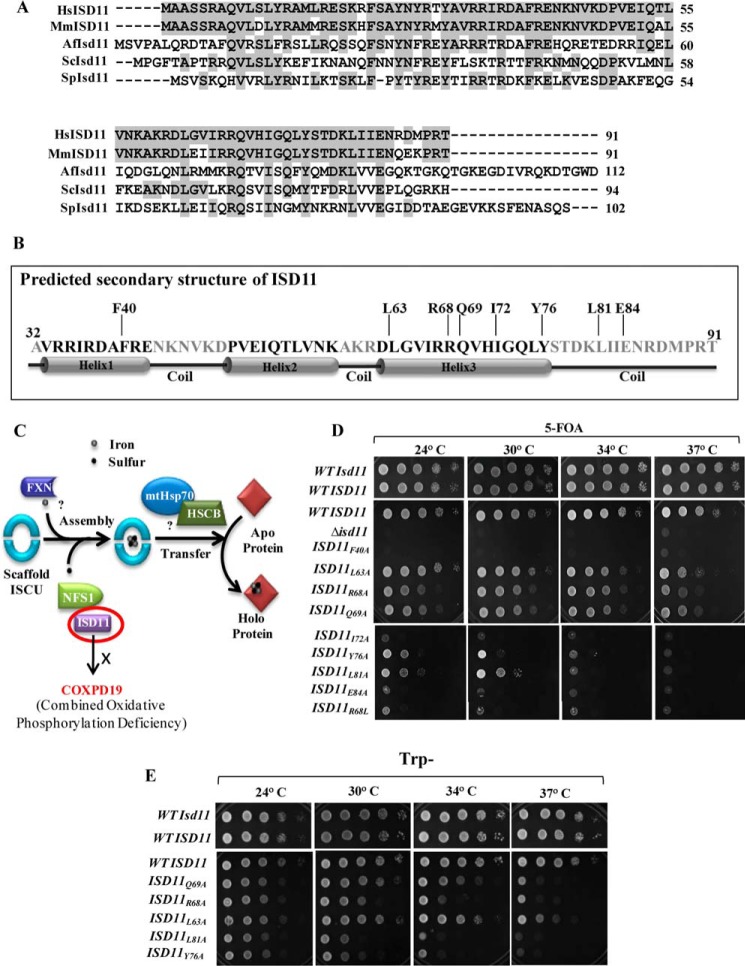
FIGURE 1.
Multiple sequence alignment of ISD11 orthologs and effect of mutations on cell viability. A, multiple sequence alignment of ISD11 protein orthologs from different species: Homo sapiens (Hs), Mus musculus (Mm), Aspergillus fumigatus (Af), S. cerevisiae (Sc), and Schizosaccharomyces pombe (Sp) was performed using ClustalW multiple sequence alignment program. Identical residues are highlighted in gray. B, secondary structure of matured form of ISD11 (amino acids 32–91) was predicted using Robetta Full-chain Protein Structure Prediction Server, depicting three helices and position of critical amino acid residues along the length of the protein. C, a schematic representation of Fe-S cluster biogenesis indicating the essential steps. Sulfur donation is assisted by NFS1-ISD11 protein complex, whereas frataxin acts as the putative iron donor. ATP-dependent mtHsp70 chaperone GRP75 and co-chaperone HSCB with the help of other transfer factors aids in the incorporation of Fe-S clusters into recipient apoproteins. R68L point mutations in ISD11 lead to development of COXPD19. D, Δisd11 yeast strain carrying a yeast WT ISD11 in pRS316 plasmid was transformed with WT LYRM4 or LYRM4 mutants (F40A, Q63A, R68A, Q69A, I72A, Y76A, L81A, E84A, and R68L) in pRS414 vector under TEF promoter and subjected to drop test analysis on 5-FOA medium followed by incubation at the indicated temperatures for 72 h. E, Δisd11 yeast strain harboring a copy of WT LYRM4 or ts LYRM4 mutants (Q69A, R68A, L63A, L81A, and Y76A) in a centromeric plasmid pRS414 TEF were subjected to spot test analysis on Trp− plates, followed by incubation for 72 h at the indicated temperatures. Δisd11 yeast strain carrying a WT ISD11 was taken as positive control.