FIGURE 5.
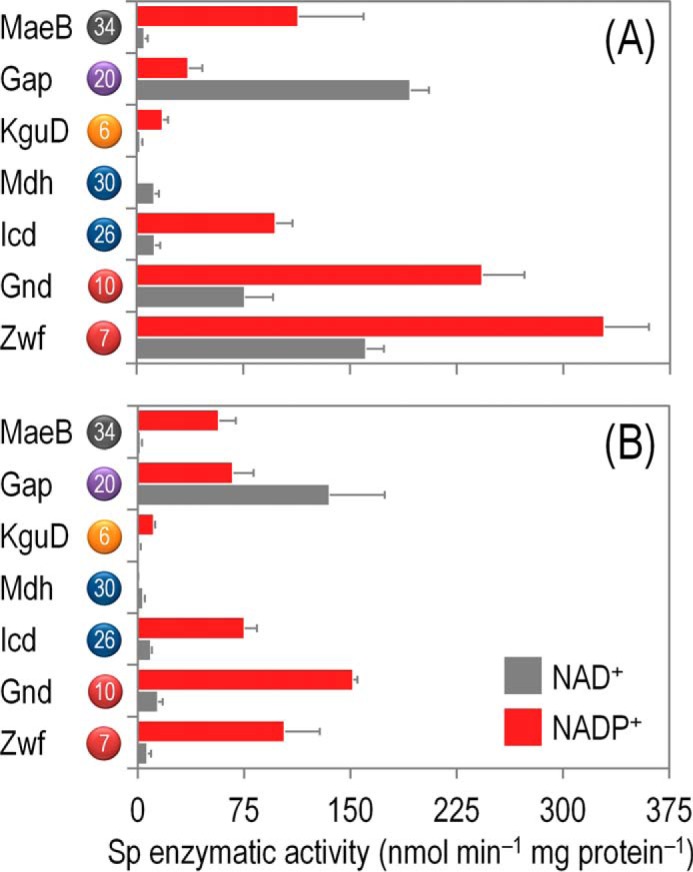
In vitro biochemical characterization of the main dehydrogenases in the core biochemical network of P. putida KT2440 under both saturating and non-saturating, quasi in vivo conditions. Specific (Sp) enzymatic activities of glucose-6-P 1-dehydrogenase (Zwf), 6-phosphogluconate dehydrogenase (Gnd), isocitrate dehydrogenase (Icd), malate dehydrogenase (Mdh), 2-ketogluconate-6-P reductase (KguD), glyceraldehyde-3-P dehydrogenase (Gap), and malic enzyme (MaeB) in cell-free extracts from P. putida KT2440 cells grown on M9 minimal medium added with 20 mm glucose during exponential growth. Assays were conducted with either saturating concentrations of substrates and cofactors (A) or in vivo-like, experimentally determined concentrations of substrates and cofactors (B). All dehydrogenases were assayed in the presence of NAD(+/H) or NADP(+/H) to assess the cofactor specificity of the corresponding enzymes (see also Table 2). Each bar represents the mean value of the corresponding enzymatic activity ± S.D. of triplicate measurements from at least two independent experiments. The circled numbers identify the enzymes in the biochemical network of Fig. 1.
