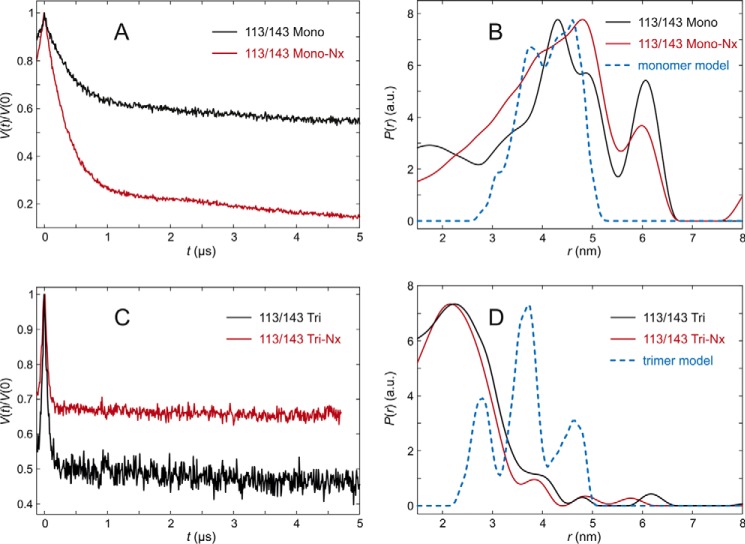
FIGURE 9.
DEER analysis for LHCII version 113/143 in n-octyl-β-d)-glucoside (OG) micelles. A, primary experimental (DEER) data in monomeric LHCII, fully pigmented (Mono, black), and lacking NX and VX (Mono-NX, red). B, distance distributions in monomeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library (blue) based on chain A only in the crystal structure (PDB code 2BHW). C, primary experimental (DEER) data in trimeric LHCII, fully pigmented (Tri), and lacking NX and VX (Tri-NX). D, distance distribution in trimeric LHCII in the presence (black) and absence (red) of NX and VX and prediction by the rotamer library based on the labels attached to chain A with chains B and C contributing to label-protein interactions (blue).