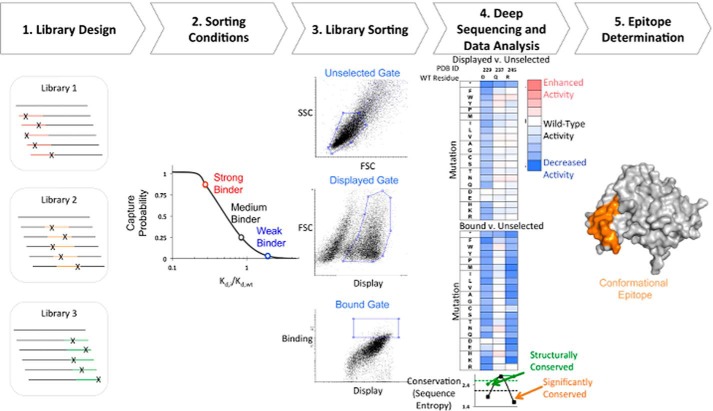
FIGURE 1.
Schematic of streamlined conformational epitope mapping process. 1, SSM libraries were made for 250–300-nucleotide contiguous sections along the gene of interest. Each library contains mutations in a different section of the gene. 2, sorting conditions were determined such that there was a higher probability of capturing stronger binding cells. 3, yeast libraries were labeled with biotinylated Fab and sorted by FACS. Three different gates are drawn: a gate on light scattering parameters SSC/FSC (top; unselected population), a gate set on FSC and fluorescence channel corresponding to display of antigen (middle; displayed population), and a binding gate that collects the top 5–10% of cells by fluorescence corresponding to channel for bound antibody (bottom; bound population). 4, DNA from each population was extracted and prepared for deep sequencing on an Illumina platform. The frequency of each variant in the bound and displayed populations is compared against the unselected population and used to calculate a fitness metric. For each residue, sequence entropy (bottom) for bound (black) and displayed (green) sorts is used to determine the degree of conservation. 5, sequence entropy is used to identify conserved and non-conserved residues that are used to determine the conformational epitope (orange).