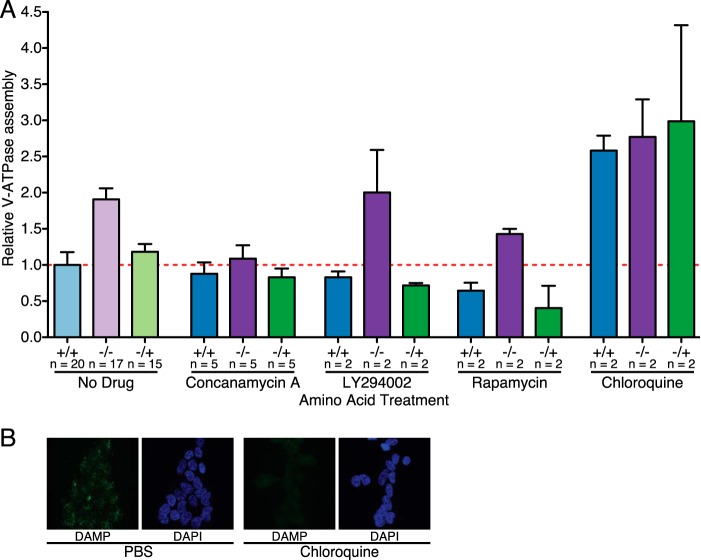
FIGURE 3.
Control of amino acid-induced changes in V-ATPase assembly. A, quantification of V-ATPase assembly in HEK293T cells maintained in amino acids (+/+), starved of amino acids (−/−), or starved of amino acids for 50 min followed by readdition of amino acids for 15-min (−/+) was performed as described for Fig. 1 except, where indicated, with the addition of 5 μm concanamycin A to inhibit the V-ATPase, 50 μm LY294002 to inhibit PI3K, 2 nm rapamycin to inhibit mTORC1, or 100 μm chloroquine to neutralize the lysosomal pH. Ratios for each set of conditions are normalized to cells maintained in amino acids (+/+) in the absence of inhibitors. p < 0.05, +/+ versus −/− and −/+. The number of independent trials is shown as n, and the error bars represent mean ± S.E. B, 100 μm chloroquine neutralizes lysosomes in HEK293T cells, as assessed by DAMP staining. HEK293T cells were grown on glass coverslips and treated with 100 μm chloroquine or medium containing an equivalent volume of PBS for 65 min. DAMP was added to the culture medium 50 min before cells were fixed to allow accumulation in acidic compartments. DAMP was detected by anti-DNP antibodies (green fluorescence), and immunocytochemistry was performed. Nuclei were stained with DAPI (blue fluorescence). Staining was assessed by confocal fluorescence microscopy.