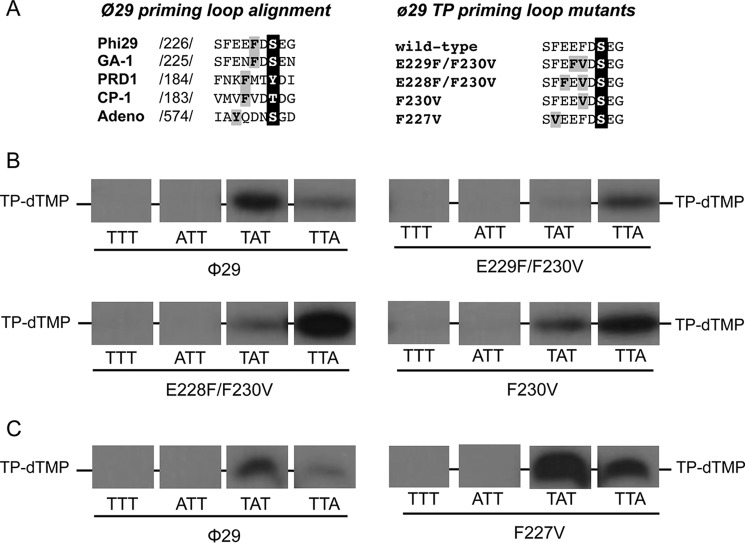
FIGURE 3.
A, alignment of the amino acid sequence of the priming loop of different viruses that initiate replication with a terminal protein. Numbers between slashes indicate the amino acid position relative to the N-terminal end of each TP. The priming residue of the different viruses is in white letters over black background. The aromatic residues are in black letters over gray background. φ29 TP priming loop mutants. The priming-loop of the wild-type and mutant TPs is shown. The primer serine is in white letters over black background and the residues changed are in black letters over gray background. B and C, ssDNA-dependent in vitro formation of the initiation complex with φ29 TP mutants of the priming loop aromatic residues Phe-230 (B) and Phe-227 (C). The assay was performed essentially as described under “Experimental Procedures” in the presence of 1 mm MnCl2. The 3′ end of the 29-mer oligonucleotide used is indicated. [α-32P]dTTP was used as initiator nucleotide. Samples were incubated for 1 h at 12 °C and then stopped, processed, and analyzed by SDS-PAGE and autoradiography. The position of the TP-dTMP complex is indicated. Panels corresponding to mutants E229F/F230V and F230V were 2-fold overexposed to better detect the initiation signal.