Table 1.
Molecules Predicted and Experimentally Confirmed To Aggregatea
| ChEMBL ID |
Structure | xLogP | AmpC IC50 no Triton (µM) |
AmpC IC50 plus Triton (µM) |
DLS detected, µM (radius, nm) |
ChEMBL ID of most similar aggregator Tc% (ECFP4 %) |
|---|---|---|---|---|---|---|
| 57997 | 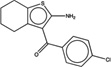 |
4.4 | 12.7 | >100 | 21 (270) |
1303983 90 (71) |
| 261275 | 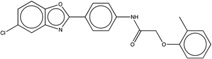 |
5.6 | 7.0 | >100 | 4 (2312) |
1329712 86 (73) |
| 1304694 | 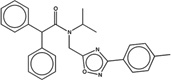 |
5.5 | 8.0 | >100 | 20 (456) |
1416661 95 (78) |
| 1333273 | 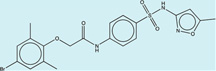 |
4.0 | 146 | >400 | 100 (49) |
1328313 89 (80) |
| 1347854 | 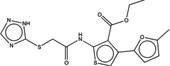 |
3.3* | 29 | >100 | 10 (115) |
1418209 85 (78) |
| 1349652 | 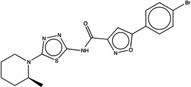 |
4.7 | 41 | >200 | 10 (484) |
6577804 96 (90) |
| 1359872 | 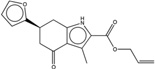 |
3.2* | 10 | >100 | 10 (405) |
326803 99 (91) |
| 1378476 | 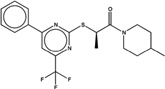 |
5.4 | 46 | >200 | 20 (163) |
1449091 96 (89) |
| 1388809 | 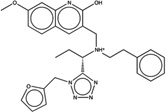 |
3.5 | 13 | >100 | 10 (177) |
1371567 90 (79) |
| 1420333 | 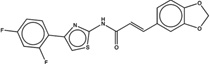 |
4.3 | 10.6 | >100 | 14 (3772) |
1310257 94 (89) |
| 1430688 | 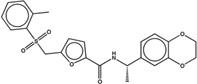 |
3.4 | 47 | >100 | 100 (240) |
1378455 86 (68) |
| 1486389 | 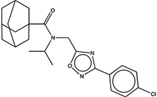 |
5.5 | 16 | >200 | 10 (248) |
1417302 96 (77) |
| 1508601 | 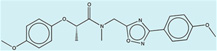 |
3.3 | 235 | >400 | 100 (62) |
1483036 91 (66) |
| 1515492 | 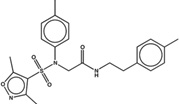 |
4.1 | 53 | >100 | 20 (142) |
1612324 92 (74) |
| 1560919 | 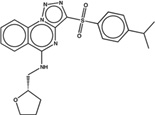 |
4.6 | 10.0 | >100 | 10 (410) |
1549087 91 (73) |
| 1565380 | 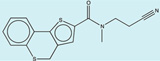 |
3.3 | 184 | >400 | 100 (71) |
1530398 91 (71) |
| 1572732 | 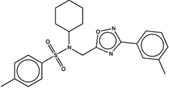 |
5.5 | 14.3 | >100 | 10 (238) |
1384931 90 (64) |
| 1607067 | 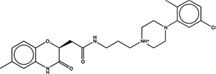 |
3.7 | 54 | >100 | 20 (291) |
1531142 90 (86) |
| 1778715 | 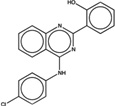 |
6.2 | 2.7 | >100 | 50 (1811) |
239009 92 (84) |
| 2165871 | 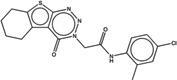 |
3.7 | 6.7 | >100 | 1.8 (2363) |
1474138 93 (85) |
| 3087539 | 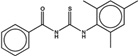 |
3.7 | 29 | >100 | 45 (383) |
1333224 93 (91) |
The criteria used to predict aggregation, xLogP and the Tanimoto coefficient to the closest known aggregators, are given, as are two experimental criteria of aggregator formation, detergent dependent inhibition of the counter-screening enzyme, AmpC, and the formation of particles detectable by DLS. Known aggregators are from refs 14 and 15.
LogP is derived from Molinspiration miLogP except those indicated by *, which are from Rdkit LogP. Weak aggregators are indicated in light blue
