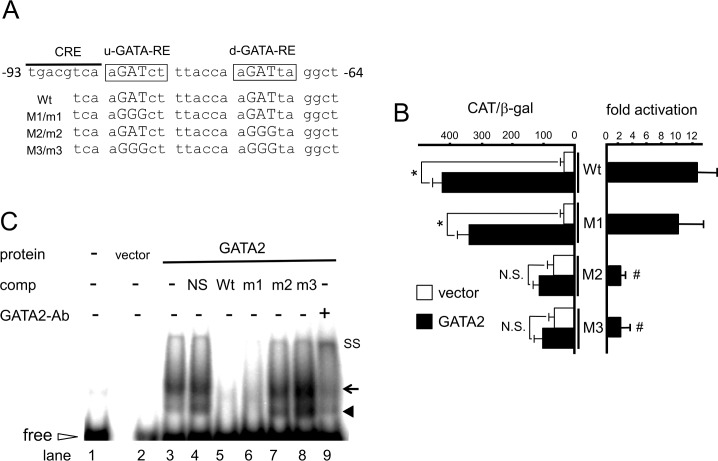
Fig 3. GATA2 recognizes u- and d-GATA-REs in the D2 promoter.
(A) A schematic representation of GATA-REs (Wt) and their mutants (M1, M2 and M3 for CAT assay; m1, m2 and m3 for gel shift assay). Two GAT sequences (open arrows) immediately downstream to CRE are designated hereafter as u- and d-GATA-RE. The sequences of wild-type (Wt) and its mutant (M1/m1, M2/m2 and M3/m3) are indicated. (B) Mutation analysis of hD2-CAT. CV1 cells were transfected with 2.0 μg hD2-CAT (Wt) or mutants (M1, M2 and M3; Fig 3A) along with 0.4 μg pcDNA3-mGATA2. Open bars, empty vector; solid bars, pcDNA3-mGATA2. CAT activity for pCMV-CAT (5.0 ng/well) was taken as 100%. Data are expressed as the mean ± S.E. of at least three independent experiments (left panel). *, P<0.05 for the empty vector vs. pcDNA3-mGATA2. To calculate fold activation (right panel), CAT activity with GATA2 was divided by that without GATA2. #, P<0.05 for hD2-CAT (Wt) vs. mutants. N.S., statistically not significant. (C) Gel shift assay using radiolabeled DNA probe containing u- and d-GATA-REs (Wt) or its mutants, m1, m2 and m3 (Fig 3A) with nuclear extract from CV1 cells transfected with pcDNA3-mGATA2. Solid arrowhead, GATA2 monomer; arrow, GATA2 dimer; open arrowhead, free probe; SS, super shift of GATA2 by the anti-GATA2 antibody.