Summary
The diarrheal pathogen Vibrio cholerae contains 3 gene clusters that encode chemotaxis-related proteins, but only cluster II appears to be required for chemotaxis. Here, we present the first characterization of V. cholerae's “cluster III” chemotaxis system. We found that cluster III proteins assemble into foci at bacterial poles, like those formed by cluster II proteins, but the two systems assemble independently and do not colocalize. Cluster III proteins are expressed in vitro during stationary phase and in conjunction with growth arrest linked to carbon starvation. This expression, as well as expression in vivo in suckling rabbits, is dependent upon RpoS. V. cholerae's CAI-1 quorum sensing (QS) system is also required for cluster III expression in stationary phase and modulates its expression in vivo, but is not required for cluster III expression in response to carbon starvation. Surprisingly, even though the CAI-1 and AI-2 QS systems are thought to feed into the same signaling pathway, the AI-2 system inhibited cluster III gene expression, revealing that the outputs of the two QS systems are not always the same. The distinctions between genetic determinants of cluster III expression in vitro and in vivo highlight the distinctive nature of the in vivo environment.
Keywords: Vibrio cholerae, chemotaxis, RpoS, quorum sensing
Introduction
One of the principal ways bacteria sense and respond to changing environmental conditions is by coupling chemotactic systems to motility, enabling them to bias their movement away from unfavorable chemical stimuli and towards favorable chemical compounds (Wadhams and Armitage, 2004; Sourjik and Armitage, 2010). Chemotactic apparatuses, which have been most fully characterized in Escherichia coli, are large, highly organized, multi-protein complexes that are generally associated with the bacterial inner membrane (Maddock and Shapiro, 1993; Sourjik and Berg, 2000). Typically, extracellular chemoeffectors are detected by the periplasmic ligand binding domains of transmembrane methyl-accepting chemotaxis proteins (MCPs). In response, MCPs modulate the activity of a phosphorelay pathway whose principal components are the cytoplasmic proteins CheW, CheA, and CheY. In E. coli (and many other organisms), phosphorylated CheY controls the flagellar switch complex that determines the direction of flagellar rotation, and thereby controls whether bacteria maintain or change their path of movement (Wadhams and Armitage, 2004; Sourjik and Armitage, 2010).
Notably, E. coli encodes only four MCPs and single copies of each of the other chemotaxis signaling proteins. In contrast, many other bacterial species encode far more MCPs and multiple homologues of chemotaxis proteins, which raises the possibility that they can sense a broader range of stimuli than E. coli, and/or respond to them in diverse ways (Szurmant and Ordal, 2004; Butler and Camilli, 2005; Hamer et al., 2010). Indeed, studies in M. xanthus, which is non-flagellated but nonetheless encodes eight chemosensory operons, have revealed a variety of outputs for chemotactic signaling pathways, including flagellum-independent chemototaxis, motility, and developmental processes (Zusman et al., 2007). Similarly, in the bacterium Rhodospirillum centenum, homologs of chemotaxis proteins have been found to mediate flagella biosynthesis and cyst development as well as classical chemotaxis (Berleman and Bauer, 2005). In other organisms, chemotaxis proteins have been found to regulate phototaxis, synthesis of Type IV pili, and several developmental processes (Zusman et al., 2007).
Vibrio cholerae, the causative agent of the diarrheal disease cholera, is a highly motile, Gram-negative rod with a single polar flagellum. Its genome encodes 68 putative chemotaxis proteins, including multiple copies of the che genes and 45 MCP-like proteins (Heidelberg et al., 2000). The mcp genes are scattered throughout the 2 chromosomes of the pathogen's genome, while the majority of the che genes are found in three clusters (I, II, and III) (Heidelberg et al., 2000; Boin et al., 2004). To date, only cluster II genes have been found to be required for chemotaxis in laboratory media (Gosink et al., 2002; Hyakutake et al., 2005). Additionally, only cluster II genes have been found to influence bacterial behavior in the suckling mouse model of infection, where they modulate the extent and localization of colonization (Butler and Camilli, 2004; Ringgaard et al., 2011; Millet et al., 2014). Both cluster I and cluster III genes have no known effect under these conditions (Millet et al., 2014). In a recent study, cluster I chemotaxis proteins were observed to localize to polar and lateral membrane regions under microaerobic conditions (standing incubation). Localization was lost upon aeration (shaking incubation), suggesting that cluster I might be involved in sensing during oxygen deprivation (Hiremath et al., 2014). However, so far no function has been observed for cluster I proteins, and it is unknown whether they contribute to chemotaxis (under specific, yet to be identified growth conditions) or instead mediate a process other than motility. Neither localization nor function has yet been reported for cluster III proteins.
In V. cholerae, unlike in E. coli, localization of the principal chemotactic signaling arrays is not a stochastic process (Ringgaard et al., 2011; Ringgaard et al., 2014). Instead, two cluster II-encoded proteins, ParC and ParP, are required for reliable localization of cluster II proteins. In new born cells, cluster II chemotaxis proteins are targeted exclusively to the old (flagellated) cell pole, whereas in pre-divisional cells, ParC and ParP recruit cluster II proteins to the new pole as well, readying this site to become an old pole after cell division (Ringgaard et al., 2011; Ringgaard et al., 2014). Mislocalization of cluster II signaling complexes results in reduced chemotactic capacity and altered swimming behavior (Ringgaard et al., 2011; Ringgaard et al., 2014). The influence of ParC and ParP on localization of cluster I and cluster III chemotaxis proteins in V. cholerae has not been reported.
We initiated the current study with the goal of defining the subcellular localization and function of the products of V. cholerae's cluster III chemotaxis-related genes. This cluster encodes all the usual components of chemotactic operons: two MCPs (one of which is predicted to be membrane associated and one that is predicted to be cytosolic), two cheWs, and one cheA, cheY, cheR, cheB and a putative cheD gene (Fig. 1A). Our efforts to link cluster III genes to chemotaxis under a variety of conditions were unsuccessful. However, we discovered that under typical laboratory culture conditions, expression of cluster III gene products is limited to stationary phase. Control of gene expression as bacteria transition into stationary phase is a complex process that is modulated by several factors, chief of which is the global stress regulator RpoS, an alternative sigma factor whose levels and activity are induced when nutrients are depleted and growth slows (reviewed in (Battesti et al., 2011; Mika and Hengge, 2014)). RpoS can also be rapidly induced in growing cells in response to a variety of different stress conditions (reviewed in (Battesti et al., 2011; Mika and Hengge, 2014)). Quorum sensing pathways, which respond to extracellular accumulation of autoinducers at high cell densities, also influence gene expression as cells enter into stationary phase (reviewed in (Ng and Bassler, 2009; Rutherford and Bassler, 2012)). V. cholerae releases two well characterized autoinducers, AI-2 and CAI-1, which are produced by the synthases LuxS and CqsA, respectively. Each autoinducer has its own receptor; however, AI-2 and CAI-I are thought to influence stationary phase gene expression via a common signaling pathway, in which the response regulator LuxO and the transcription regulator HapR play pivotal roles (reviewed in (Ng and Bassler, 2009; Rutherford and Bassler, 2012)).
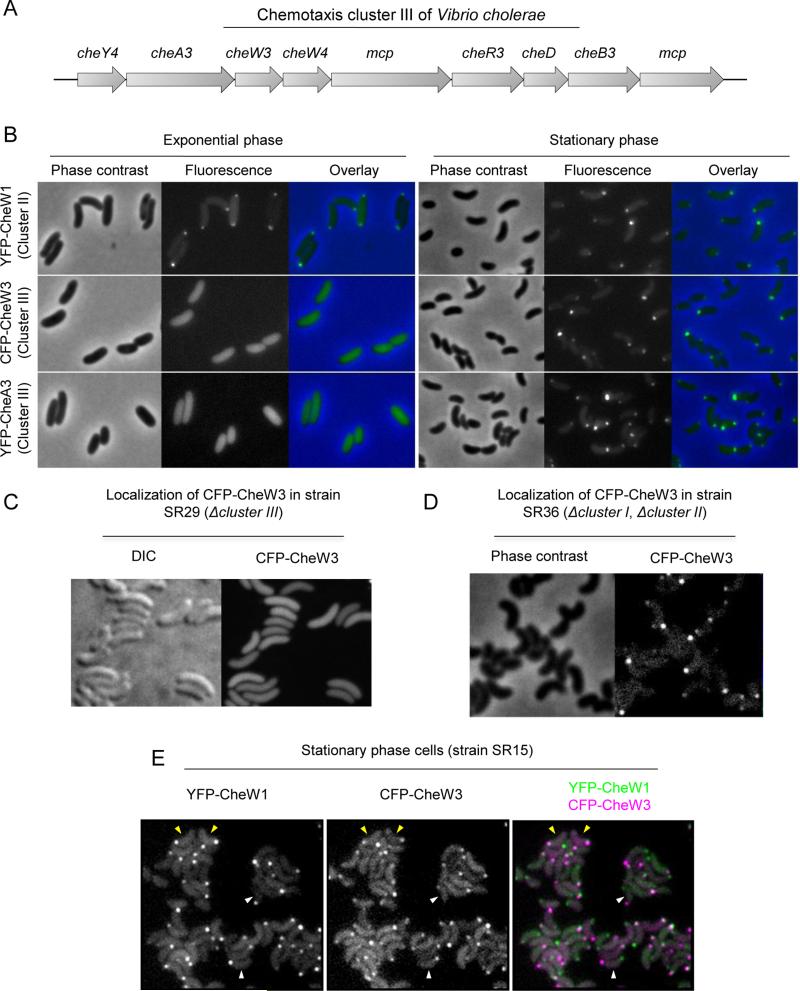
Figure 1. Growth phase dependent polar localization of chemotaxis cluster III proteins.
(A) Schematic of chemotaxis cluster III. (B) Localization of ectopically expressed YFP-CheW1, CFP-CheW3, and YFP-CheA3 in cells from exponential (left) and stationary (right) phase cultures of wild type V. cholerae. (C, D) Localization of CFP-CheW3 in cluster III-deficient strain SR29 (C) and in SR36, a strain deleted for chemotaxis clusters I and II (D). Cells were grown to stationary phase. (E) Localization of YFP-CheW1 and CFP-CheW3 expressed from their respective native loci (in strain SR15) in stationary phase cells. Yellow arrowheads indicate cells with YFP-CheW1 and CFP-CheW3 localized to the same pole. White arrowheads indicate cells where YFP-CheW1 and CFP-CheW3 localize to opposite poles.
We have described environmental as well as genetic factors that govern expression of V. cholerae cluster III proteins, as well as explored their subcellular distribution. As noted above, expression of cluster III proteins (unlike cluster II proteins, which are constitutively expressed) is induced by entry into stationary phase, and can also be induced by carbon starvation. Expression is positively regulated by RpoS (both in response to growth phase and carbon starvation) and CqsA (growth phase only), but unexpectedly appears to be repressed by LuxS, countering the idea that these QS pathways have a single output. As with cluster II proteins, cluster III proteins form polar foci; however, they are not restricted to the old pole, consistent with their lack of a role in flagellum-mediated motility. Notably, we observed expression and polar localization of cluster III proteins during V. cholerae infection of infant rabbits. Localization, and most likely expression, of cluster III proteins in vivo required RpoS but was in part independent of CqsA. The distinct requirements for expression/localization of cluster III proteins in vivo and in vitro highlight the unusual physiologic state of V. cholerae during infection.
Results
Subcellular localization of chemotaxis cluster III proteins is growth phase dependent
We previously found that chemotaxis proteins encoded in V. cholerae chemotaxis cluster II localize to the cell pole in a cell cycle dependent manner (Ringgaard et al., 2011; Ringgaard et al., 2014). To investigate if proteins encoded in chemotaxis cluster III display a similar localization pattern, we compared the localization of ectopically expressed YFP-CheA3 and CFP-CheW3, translational fusions of two cluster III-encoded proteins and fluorescent proteins, with that of YFP-CheW1, a fluorescently tagged cluster II-encoded protein. In contrast to YFP-CheW1, which localized to the cell poles in both exponentially growing and stationary phase cells, we observed only a diffuse cytoplasmic signal from CFP-CheW3 and YFP-CheA3 in exponential phase cells (Fig. 1B). However, strikingly, in stationary phase cells, both cluster III-encoded proteins localized to the cell poles (Fig. 1B, right panels). These observations suggest that the polar localization of cluster III proteins depends on a factor that is only produced (or active) in stationary phase.
We also explored whether the polar localization of CFP-CheW3 was dependent on additional chemotaxis proteins. First, to test whether CheW3 localization required additional cluster III gene products, we imaged ectopically expressed CFP-CheW3 in SR29, a strain lacking chemotaxis cluster III. Polar CFP-CheW3 foci were not observed in stationary phase cells in this background (Fig 1C), suggesting that interactions between cluster III proteins are required for generating detectable CheW3 foci. Since MCPs are required for array formation in other organisms, it is likely to be the absence of the cluster III MCPs that leads to diffuse CFP-CheW3 localization in the cluster III mutant background. In contrast, when we imaged CFP-CheW3 in a strain lacking chemotaxis clusters I and II (SR36), we found that CFP-CheW3 localized to the poles (Fig. 1D). Thus, polar localization of cluster III proteins is independent of the other two chemotaxis clusters; there is no apparent crosstalk between cluster III localization and the ParC/ParP-mediated polar localization of cluster II proteins (Ringgaard et al., 2011; Ringgaard et al., 2014).
Consistent with the idea that cluster II and cluster III proteins form independent complexes, simultaneous visualization of cluster II and III proteins (in strain SR15) revealed that YFP-CheW1 and CFP-CheW3 did not always localize to the same pole (Fig. 1E, white arrowheads). Furthermore, even when both proteins were found at the same pole (Fig. 1E, yellow arrowheads), foci of YFP-CheW1 and CFP-CheW3 did not always perfectly co-localize. Thus, although cluster III proteins, like those of cluster II, appear to form macromolecular signaling complexes at cell poles, there is no evidence for crosstalk between these complexes or any shared function.
Cluster III proteins are only expressed in stationary phase
To further explore the growth phase dependent localization of CheW3, we used a strain (SR9) that contains CFP-CheW3 expressed from its native chromosomal site under the control of its native promoter. When overnight cultures (late stationary phase) of SR9 were diluted into fresh LB medium, ~65% of stationary phase cells (at time 0) had a polar CFP-CheW3 focus (Fig. 2A, B). Afterwards, the percentage of cells with a CFP-CheW3 focus decreased as the cells started growing, until no cells were observed to possess CFP-CheW3 foci at the onset of exponential growth. As cultures approached their maximal density (OD600 ~ 3.5), polar CFP-CheW3 foci reappeared, and within one generation reached the starting level of ~65% of cells with polar CFP-CheW3 foci (Fig. 2B). A similar growth phase-dependent localization pattern was observed when cells were grown in M9 minimal medium supplemented with thiamine and glycerol. However, in minimal medium nearly 100% of cells possessed CFP-CheW3 foci in stationary phase (Fig. 2C and Fig. S1).
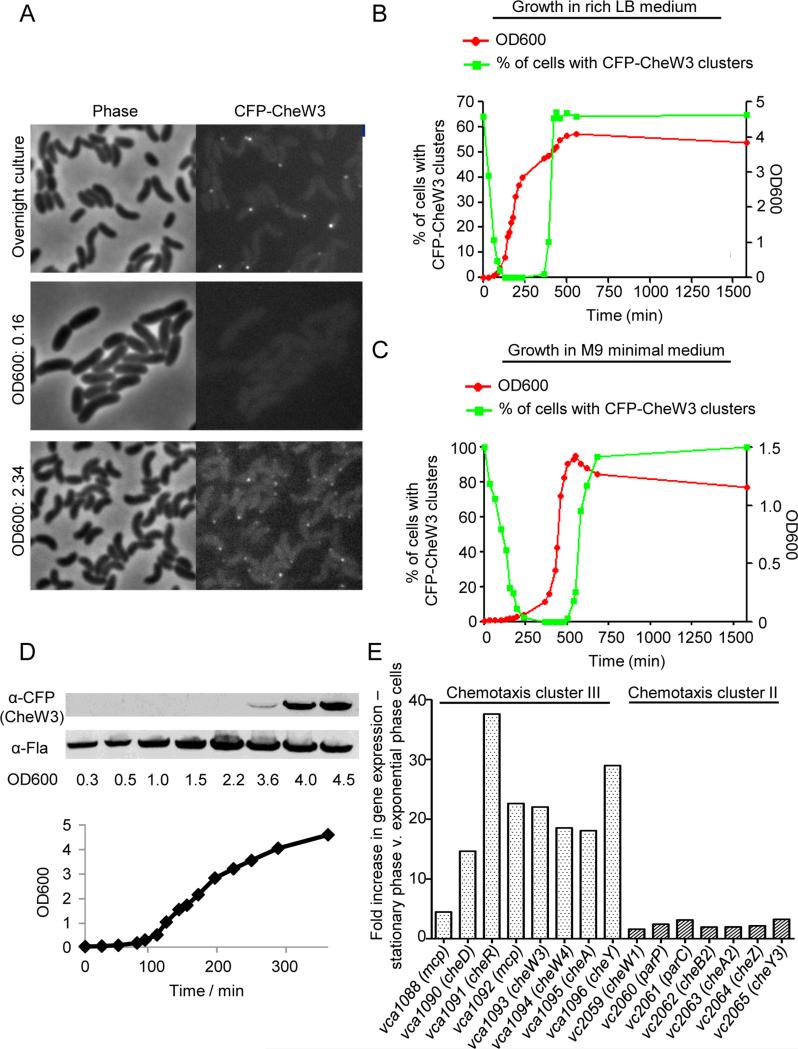
Figure 2. Correlation between stationary phase specific polar localization and expression of chemotaxis cluster III proteins.
(A) Localization of CFP-CheW3 expressed from its native locus and promoter (in strain SR9) at different optical densities during growth in LB. (B) Growth curve (filled green squares) of strain SR9 in LB medium and the corresponding percentage of cells with CFP-CheW3 foci (filled red circles). (C) Growth curve (filled green squares) of strain SR9 in minimal M9 medium and the corresponding percentage of cells with CFP-CheW3 foci (filled red circle). See Figure S1 for micrographs. (D) Western blot against CFP-CheW3 and FlaA using anti-CFP and anti-FlaA specific antibodies on SR9 cultures from the indicated optical densities. Samples were normalized and loaded with equal optical density. The corresponding growth curve is shown. (E) RNAseq experiment of the transcription levels of cluster II and III chemotaxis genes in stationary and exponentially growing cells. The Bar-graph shows the fold change in gene expression between stationary phase cells versus exponentially growing cells.
We also monitored CFP-CheW3 expression in different growth phases by western blotting with antisera to CFP (Fig. 2D). There was an excellent correlation between detection of CFP-CheW3 foci and of CFP-CheW3 by western. No CFP-CheW3 was detected in exponentially growing cells, and the protein became detectable when the culture reached stationary phase (OD600 ~ 3.6). We used expression of FlaA, the major V. cholerae flagellin, to control for the total protein in the samples from the different time points (Fig. 2D). Although there was somewhat less FlaA apparent at the early time points, the differences cannot account for the marked increase in CFP-CheW3 detected during stationary phase. Since cluster III genes are thought to be expressed as an operon, and since formation of CFP-CheW3 foci is dependent upon the presence of cluster III, this result suggests that additional proteins of cluster III are also induced as cultures become saturated. RNAseq analyses provided additional evidence that numerous cluster III genes, including cheW3, are more highly expressed in stationary phase than in exponential phase growth (Fig. 2E). Markedly more transcripts (5 – 35 fold more) for eight cluster III genes were detected in stationary phase RNA. In contrast, we observed only a minor increase in cluster II gene expression upon entry into stationary phase (Fig. 2E). Thus, the increase in CheW3 upon entry into stationary phase is at least in part due to changes in transcriptional regulation that is specific to cluster III chemotaxis genes.
RpoS and quorum sensing regulate expression of chemotaxis cluster III genes in stationary phase cells
Since the stationary phase sigma factor RpoS and quorum sensing pathways are key modulators of gene expression in stationary phase, CFP-CheW3 reporter strains lacking RpoS or auto inducer synthases CqsA (CAI-1 synthesis) or LuxS (AI-2 synthesis) were constructed to address whether these factors influence cluster III gene expression. In marked contrast to wild type stationary phase LB cultures, which had polar CFP-CheW3 foci in ~65% of cells, the rpoS and the cqsA mutants (strains SR71 and SR48, respectively) completely lacked CFP-CheW3 foci (Fig. 3A, B). However, a higher percentage of ΔluxS cells (strain SR49) than wild type cells contained polar CFP-CheW3 foci (~90%), and foci were brighter than in the wild type background (Fig. S2). Thus, RpoS and CqsA are required for formation of CFP-CheW3 foci in stationary phase cells, while LuxS appears to inhibit focus formation. The ΔcqsAΔluxS double mutant (strain SR47) completely lacked CFP-CheW3 foci, indicating that the CqsA-mediated phenotype is dominant over the LuxS phenotype regarding formation of CFP-CheW3 foci in stationary phase cultures.
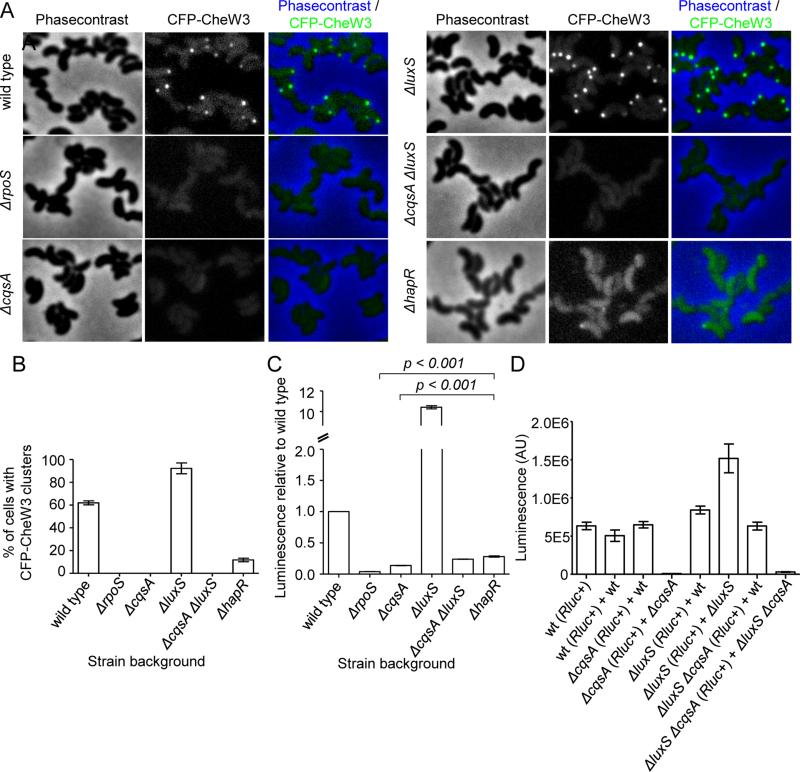
Figure 3. RpoS and quorum sensing signaling modulate expression of CheW3.
(A) Visualization of CFP-CheW3 expressed from its native locus in late stationary phase wild type (strain SR9) cells and in various mutant derivatives: ΔrpoS (strain SR71), ΔcqsA (strain SR48), ΔluxS (strain SR49), ΔcqsAΔluxS (strain SR47), ΔhapR (strain SR12). (B) Bar-graph showing the percentage of cells with CFP-CheW3 foci in strain SR9 and in various mutant derivatives: ΔrpoS (strain SR71), ΔcqsA (strain SR48), ΔluxS (strain SR49), ΔcqsAΔluxS (strain SR47), ΔhapR (strain SR12). (C) Total luminescence of the mutant strains ΔrpoS (strain SR84), ΔcqsA (strain SR81), ΔluxS (strain SR82), ΔcqsAΔluxS (strain SR80), ΔhapR (strain SR89) relative to wild type V. cholerae strain SR76 (Rluc8-CheW3). All cultures were grown in LB medium. (D) Total luminescence of stationary phase cells from co-cultures of cells. Wild type (SR76), ΔcqsA (SR81), ΔluxS (SR82), and ΔcqsAluxS (SR80) rluc-cheW3+ strains were grown in co-cultures with rluc-cheW3− wild type (C6706), ΔcqsA (SR48), ΔluxS (SR49), and ΔcqsAluxS (SR47) in order to test if production and release of AI-1 and AI-2 from wild type could complement expression (luminescence) of cluster III in mutants.
We constructed a translational fusion between full-length CheW3 and Renilla reniformis luciferase (Rluc) expressed from the native cheW3 locus to address if changes in expression of CheW3 (monitored with luminescence) could account for the differences in the frequency of CheW3 foci observed above. In these experiments, expression of rluc-cheW3 in the ΔrpoS, ΔcqsA, ΔluxS, and ΔcqsAΔluxS backgrounds was compared to that in the wild type, which was set at 1 (Fig. 3C). There was very little expression of Rluc-CheW3 observed in ΔrpoS, ΔcqsA, and ΔcqsAΔluxS strains, suggesting that the absence of CFP-CheW3 foci in these background is due to lack of CheW3 expression. In contrast, there was a greater than tenfold increase in expression of Rluc-CheW3 in the ΔluxS mutant, suggesting that the increased frequency of focus formation and fluorescence intensity in this background are due to elevated levels of CFP-CheW3 and other cluster III proteins. Both the reduced reporter activity in the ΔcqsA and ΔcqsAΔluxS strains and the elevated reporter activity in the ΔluxS strain could be restored to wild type levels by co-culture with wild type cells, presumably due to provision of autoinducers in trans (Fig. 3D). Collectively, these observations suggest that, in stationary phase cells, RpoS and CqsA (via CAI-1) promote expression of CheW3 (and likely other chemotaxis cluster III proteins), whereas LuxS (via AI-2) inhibits its expression. The opposite effects of LuxS and CqsA on expression of cluster III genes in stationary phase was unexpected, since their autoinducer products are both thought to feed into the same regulatory pathway, ultimately leading to dephosphorylation of LuxO and increased expression of HapR at high cell density (Ng and Bassler, 2009; Rutherford and Bassler, 2012).
To further decipher this regulatory process, we investigated cluster III expression and focus formation in a HapR-deficient strain. In a ΔhapR mutant (strain SR12), we did occasionally observe polar localization of CFP-CheW3 and cells appeared to have a more intense cytosolic CFP signal (Fig. 3A); however, the percentage of cells with foci was much reduced compared to wild type (Fig. 3B). Furthermore, expression of Rluc-CheW3 was much lower in the ΔhapR mutant than in the wild type, although significantly higher than that of the ΔcqsA and the ΔcqsAΔluxS mutants (Fig. 3C), which is consistent with the microscopy data of CFP-CheW3 in the ΔhapR mutant. This finding, coupled with our earlier observations, suggests that cluster III genes are downstream of HapR in the V. cholerae quorum sensing pathway, and that the CqsA-synthesized CAI-1 is the dominant activator of HapR expression in these cultures. A more prominent role for CqsA than LuxS has also been observed in analyses of quorum-regulated biofilm formation (Zhu and Mekalanos, 2003). However, it is not clear why the LuxS-deficient strain shows enhanced expression of cluster III genes and production of cluster III foci. We note that cluster III foci were still detected in ~10% of stationary phase HapR-deficient cells (Fig. 3B), and that there is some HapR-independent expression of cluster III genes (Fig. 3C). Thus, it is possible that LuxS/AI-2 exert a negative regulatory effect on a HapR-independent pathway, which enables elevated cluster III expression in the luxS mutant.
Carbon starvation induces expression and localization of chemotaxis cluster III proteins
Our observation that a higher percentage of cells possess CheW3 foci in minimal medium (~100%, Fig. 2C, S1) than in LB (~65%) (Fig. 2AB) suggested that starvation could also be a factor modulating expression of chemotaxis cluster III genes. The Rluc-CheW3 luciferase reporter strain was used to monitor CheW3 expression when cells were depleted for carbon sources (Fig. 4A). In these experiments, overnight cultures of a wild type strain harboring the rluc-cheW3 reporter (SR76) were used to inoculate fresh MSR6 minimal medium supplemented with 0.01% to 0.4% (w/v) D-glucose. Cells were grown in microtiter plates and the OD600 and total luminescence of each well was measured as a function of time. A concentration of 0.4% glucose was sufficient for cells to grow into saturated stationary phase cultures, while at lower concentrations bacterial growth arrested at lower culture densities, presumably due to exhaustion of D-glucose (Fig. 4A). There was a clear correlation between the time when growth ceased and markedly increased expression of Rluc-CheW3 (Fig. 4A), such that elevated CheW3 expression was detected first in cultures with 0.01% glucose and last in cultures that contained 0.4% glucose (Fig 4A). Thus, growth arrest linked to carbon starvation is a potent inducer of CheW3 expression, even when cells are not grown to high density.
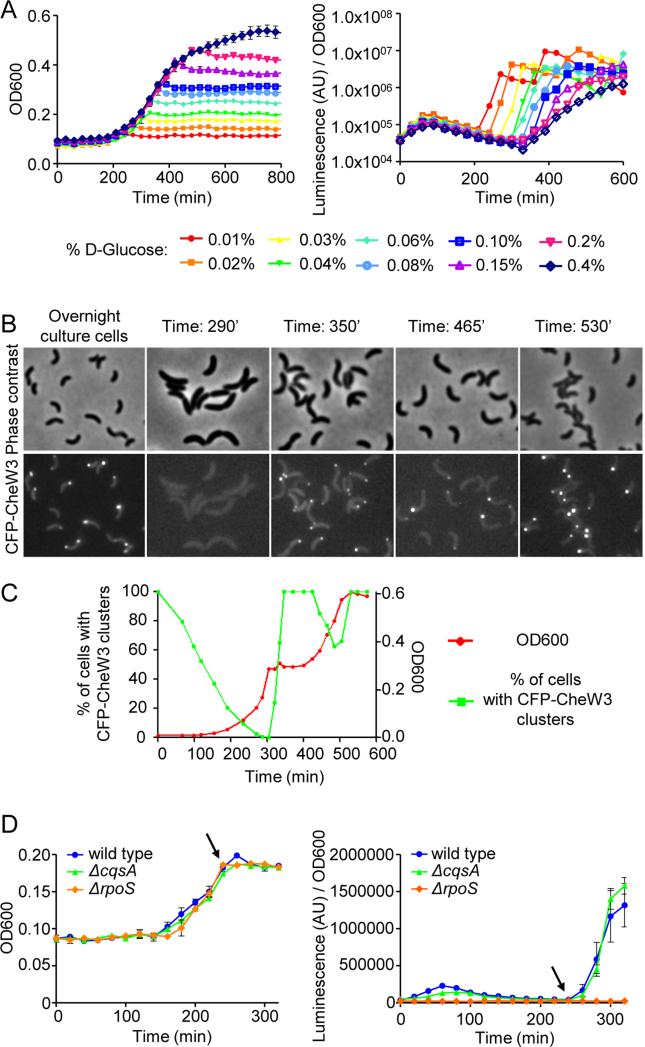
Figure 4. Growth arrest following carbon starvation induces expression/localization of CheW3 in an RpoS dependent and CqsA independent fashion.
(A) Optical density (OD600) and total luminescence were measured over time for wild type V. cholerae strain SR76 (Rluc8-CheW3). Cells were grown in MSR6 minimal medium with the indicated concentrations of D-glucose. (B) Detection of CFP-CheW3 foci at different time points during the carbon catabolite experiment shown in (C). (C) Growth-curve (filled green squares) of strain SR9 (cfp-cheW3) in MSR6 minimal medium supplemented with 0.04% w/v D-glucose and 0.4% w/v succinate and the corresponding percentage of cells with CFP-CheW3 foci (filled red circle) at the indicated optical densities. (D) Kinetics of CheW3 expression, detected using Rluc8-CheW3 luminescence, as a function of growth in wild type V. cholerae (strain SR76), ΔrpoS (strain SR84) and ΔcqsA (strain SR81) backgrounds.
To further explore the relationship between CheW3 expression/focus formation and carbon metabolism/cell growth, we set up a carbon catabolite repression assay (Madigan, MT, Martinko, JM, Clark, 2000). Cells were grown in a very minimal medium (MSR6) supplemented with 0.04% w/v D-glucose and 0.4% w/v succinate. In these conditions, the cells initially only metabolize D-glucose; when it is used up, they arrest growth temporarily, then resume growth by metabolizing succinate. An overnight culture (late stationary phase) was diluted into fresh MSR6 medium, and growth and focus formation was monitored as in Fig. 2C (Fig. 4B-C). As observed in M9 minimal medium, 100% of stationary phase cells had a polar CFP-CheW3 focus in MSR6 medium. The percentage of cells with a CFP-CheW3 focus decreased as the cells progressed through lag phase, so that by the onset of exponential growth no cells with CFP-CheW3 foci were observed. The subsequent growth arrest (at OD~0.3), indicative of glucose depletion, was rapidly followed by an increase in the percentage of cells with CFP-CheW3 foci; within 20 minutes of arrest, 100% of cells had polar CFP-CheW3 foci (Fig. 4B-C). When cell growth resumed, presumably as cells started metabolizing succinate (at ~400 minutes), there was a concomitant decrease in the percentage of cells with CFP-CheW3 foci. Finally, as the culture entered stationary phase (after 500 minutes), the percentage of cells with CFP-CheW3 foci increased again to 100% (Fig. 4B-C). Together, these observations illustrate the remarkable correlation between growth arrest (caused either by carbon starvation or by culture saturation) and induction of expression and polar localization of V. cholerae chemotaxis cluster III proteins.
We used the Rluc-CheW3 based luminescence assay to test whether RpoS or CqsA are required for the expression of cluster III genes that follows growth arrest induced by carbon starvation (Fig. 4D). When wild type, ΔcqsA, and ΔrpoS strains were grown in MSR6 minimal medium supplemented with 0.04% w/v D-glucose (Fig 4D left, arrow), expression of CheW3 increased rapidly after growth ceased in the wild type and the ΔcqsA backgrounds (Fig. 4D, arrow). However, no luminescence was detected in the ΔrpoS background, indicating that RpoS, but not CqsA, is absolutely required for expression of cluster III proteins following growth arrest induced by carbon starvation. Thus, the requirements for expression and localization of cluster III proteins differ between stationary phase LB, where both RpoS and CqsA are necessary, and growth arrest due to carbon starvation, for which only RpoS is necessary (Fig. 3).
Since carbon starvation induces cluster III expression, we analyzed if a cluster III mutant would be out-competed by the wild type in competition experiments when cells were starved for carbon over longer periods. Wild type (lacZ+) and cluster III mutant (lacZ−) were co-inoculated 1:1 in MSR6 minimal supplemented with 0.04% w/v L-glucose (which induces growth arrest due to carbon limitation during exponential growth), cells were incubated over night, and the ratio between wild type and mutant was determined by plating on X-gal indicator plates. Additionally, the culture was back-diluted into three different growth conditions: MSR6 minimal supplemented with 0.04% w/v L-glucose, MSR6 minimal supplemented with 0.04% w/v L-glucose, and LB rich medium and incubated over night. This cycle continued for seven days and each day the ratio between wild-type and mutant was determined. We observed no effect of the absence of cluster III (Fig. S3) when cells were starved for carbon. Furthermore, when we performed chemotactic assays with stationary phase cells (expressing cluster III), we observed no chemotactic defect of cluster III mutants when compared to wild type. As expected, a cluster II mutant was outcompeted by wild type in chemotactic assays (Fig. S4).
Both RpoS and CqsA contribute to expression/polar localization of CheW3 during V. cholerae intestinal colonization
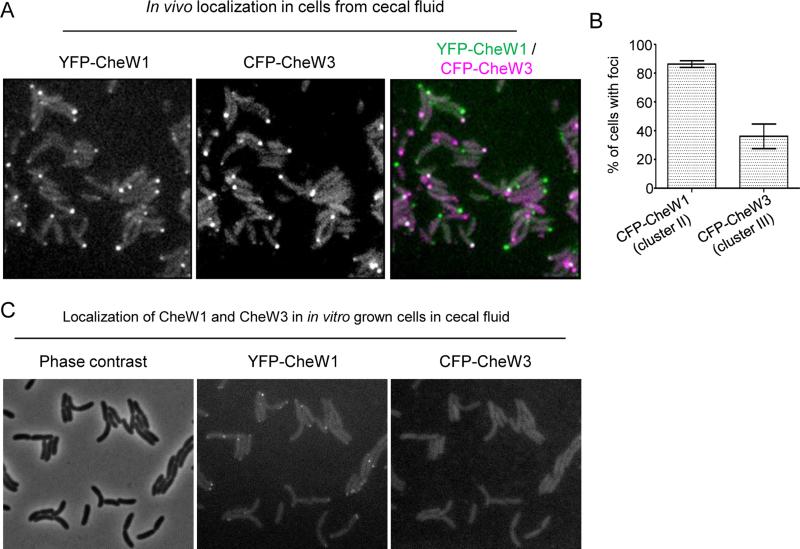
Infant rabbits infected with V. cholerae develop severe cholera-like diarrhea (Ritchie et al., 2010), and we found it is possible to image the intracellular localization of fluorescently tagged V. cholerae proteins in cells collected from cecal fluid, in which they accumulate to densities of >109 cfu/ml. Rabbits were inoculated with the YFP-CheW1 and CFP-CheW3-producing strain SR15, which colonizes and causes disease equivalent to its wild type parental strain C6706 (Fig. S5, (Ritchie et al., 2010)). Notably, nearly 90% of intestinal bacteria contained polar foci of YFP-CheW1 (a cluster II protein), and ~35% of cells had detectable polar foci of the cluster III protein CFP-CheW3 (Fig. 5A, 5B). These observations demonstrate that cluster III genes are expressed during infection, corroborating previous RNA-Seq and microarray based analyses (Nielsen et al., 2006; Mandlik et al., 2011), and show for the first time that it is possible to monitor the subcellular localization of bacterial proteins during infection. We also observed polar localization of cluster III proteins in homogenized tissue from the distal small intestine of infected rabbits (Fig. S6), although quantification of foci in these samples was not possible due the presence of autofluorescent tissue residues in these samples. However, when SR15 cells were taken from exponentially growing LB cultures and inoculated into filtered (cell-free) cecal fluid, CFP-CheW3 foci were not detected after 2hrs of growth at 37° (Fig. 5C). This observation suggests that V. cholerae survival, growth and passage through the small bowel may be required to induce cluster III genes, rather than simply the presence of an inducing signal within cecal fluid. It is also noteworthy that cells continued to grow and divide during the 2hr observation period, suggesting that cecal fluid is not a nutrient-limited condition, and thus that in vivo expression of cluster III genes, unlike in vitro expression, may not be association with cessation of growth.
Figure 5. Intracellular localization of YFP-CheW1 and CFP-CheW3 in cells from cecal fluid during V. cholerae colonization of infant rabbits.
(A) Fluorescent micrographs and (B) percentage of cells from cecal fluid with YFP-CheW1 and CFP-CheW3 clusters from cecal fluid infant rabbits infected with V. cholerae strain SR15 (YFP-CheW1, CFP-CheW3). (C) Localization of YFP-CheW1 and CFP-CheW3 in SR15 cells initially grown to exponential phase in LB and then transferred to filtered cecal fluid an imaged after 2hr incubation at 37° with shaking.
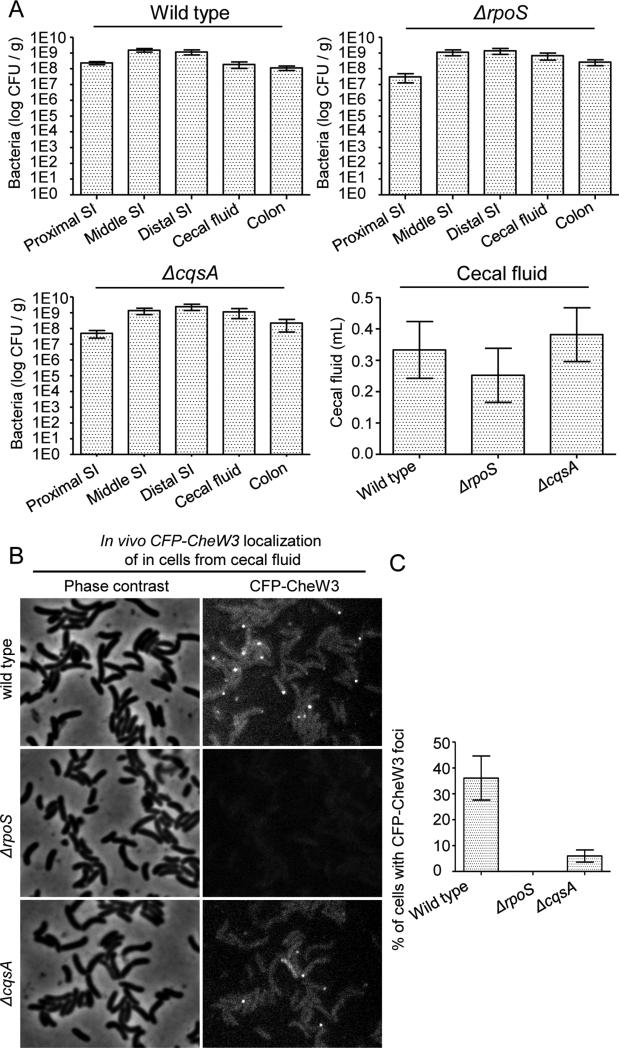
We used CFP-CheW3 foci as a marker to explore the in vivo requirements for expression of cluster III proteins. In particular, we tested whether RpoS and CqsA, which are required for stationary phase expression of cluster III genes in vitro (Fig. 3), are also required for in vivo expression/localization of CFP-CheW3. Mutants carrying a chromosomal cfp-cheW3 reporter and lacking rpoS or cqsA (SR71 and SR48, respectively) colonized the rabbit intestine and induced cecal fluid accumulation equivalent to wild type V. cholerae (strain SR9) (Fig. 6A). However, we did not detect any CFP-CheW3 foci in RpoS-deficient cells taken from cecal fluid of infected rabbits (Fig. 6B,C). These observations indicate that rpoS is essential for CheW3 expression/focus formation in vivo, and confirm that expression of cluster III genes is not important for V. cholerae pathogenicity, as previously reported for infection of infant mice, where deletion of cluster III has no influence on colonization (Millet et al., 2014). In contrast to rpoS, cqsA was not required for CFP-CheW3 expression/localization in cells from cecal fluid. However, the percentage of cells with foci was reduced compared to that of wild type cells (Fig. 6B, C), suggesting that CqsA also contributes to expression of cluster III in vivo. The different genetic requirements for CheW3 expression in the gut compared to in vitro cultures suggest that there may be multiple stimuli/pathways that can lead to cluster III induction.
Figure 6. RpoS is required for expression/localization of CheW3 during V. cholerae colonization of the infant rabbit intestine.
(A) Number of colony forming units (CFUs) per gram from homogenates of the indicated sections of the small intestine (SI), colon or the cecal fluid. Cecal fluid accumulation in rabbits infected with wild type (SR9), SR71 (ΔrpoS, cfp-cheW3), or SR48 (ΔcqsA, cfp-cheW3) V. cholerae. (B) Fluorescent micrographs and (C) percentages of cells from cecal fluid with CFP-CheW3 foci from infant rabbits infected with SR9, SR76, and SR48.
Discussion
Many bacterial species encode several independent chemotaxis operons, suggesting that their chemotaxis pathways are much more complex than that of the model organism E. coli. However, the regulation and purpose of multiple chemotaxis systems has only been studied in a few bacterial species. We show here that the in vitro expression of V. cholerae chemotaxis cluster III proteins coincides with cessation of growth, either due to carbon starvation or to culture saturation, and is dependent in both circumstances upon RpoS. The CAI-1 and AI-2 quorum sensing pathways were also found to regulate cluster III gene expression, although in opposite direction, which counters prevailing dogma. Visualization of fluorescently tagged proteins revealed that cluster III genes are expressed in a subset of V. cholerae in vivo, potentially independently of growth cessation. As for cluster II chemotaxis proteins, foci of cluster III chemotaxis proteins were observed at cell poles, both in vitro and in vivo; however, the two systems appears to localize independently. Our data highlight the diversity of stimuli and regulatory processes that govern expression of these chemosensory proteins.
Our work provides the first demonstration that cluster III chemotaxis proteins, like those of clusters I and II, assemble into cellular foci, as well as the first visualization of chemotactic signaling arrays during infection. Cluster III foci form at bacterial poles; however, they do not precisely colocalize with cluster II foci and in some cells are instead at the opposite pole. Their distinction from the flagellum-associated cluster II foci suggests that this putative chemosensory apparatus may have an output other than flagellar motility. Further supporting this idea, despite intensive efforts using cells grown under cluster III-inducing conditions, we have been unable to detect any chemotaxis-related phenotype associated with the presence or absence of cluster III genes. Diverse outputs from chemosensory pathways have been identified in other organisms that contain multiple chemotaxis gene clusters (Berleman and Bauer, 2005; Zusman et al., 2007); however, additional work is needed to define the targets of V. cholerae's cluster III proteins.
Expression of cluster III genes is governed by a variety of regulatory processes. Previous microarray-based analyses of mRNA levels have demonstrated that expression of cluster III genes increases at stationary phase in vitro and during growth within ligated intestinal loops from adult rabbits, and that both HapR and RpoS contribute to this increase (Nielsen et al., 2006). However, earlier analyses have not reported whether gene expression correlated with the presence of chemotactic proteins and protein complexes or was detectable during “uninduced” conditions, nor explored the influence of quorum sensing. Our analyses suggest that the dominant regulatory factor may be the alternative sigma factor RpoS, as it is required for expression under all conditions tested. However, the specific carbon-starvation signal that triggers the RpoS-dependent expression of cluster III remains to be elucidated. It is, however, likely that cluster III is part of the RpoS-mediated general stress response, which leads to general stress resistance of cells upon nutrient deprivation, stress or entry into stationary phase (Battesti et al., 2011). During exponential growth RpoS translation is inhibited and RpoS protein is rapidly degraded, resulting in a very low pool of RpoS. The RpoS response is induced by a rapid increase in levels of RpoS and is regulated at both the transcriptional and translational levels, and through degradation, and regulation of RpoS activity.
Expression of RpoS is regulated by a variety of signals, including (at the transcriptional level) HapR (Joelsson et al., 2007). Since the autoinducer synthase CqsA (whose product induces HapR expression) is also required for most or all expression of the cluster III gene CheW3 in stationary phase cultures, it is possible that the key role of CqsA (with respect to cluster III) is induction of RpoS. However, CqsA is not required for cluster III gene expression in response to carbon starvation, and it contributes to but is not essential for cluster III gene expression in vivo. The fact that CqsA modulates cluster III expression in vivo suggests that intraintestinal V. cholerae cell densities are sufficient to activate quorum sensing pathways, at least in a subset of cells, and that carbon starvation cannot be the only stimulus for in vivo induction of cluster III genes. Furthermore, since cecal fluid can support continued V. cholerae replication, it seems likely that stimuli not identified here (i.e., neither carbon starvation nor quorum sensing) also contribute to RpoS induction in vivo.
Given the role we have described for CqsA, as well as previously reported overlapping roles for CqsA and V. cholerae's second autoinducer synthase, LuxS (Ng and Bassler, 2009; Rutherford and Bassler, 2012), our observation that LuxS is a negative regulator of cluster III expression is unexpected. The products of both CqsA and LuxS are thought to signal through the same LuxO- and HapR-mediated pathway, although previous analyses indicate that CqsA provides a more powerful signal (Miller et al., 2002; Joelsson et al., 2007). However, our data suggests that LuxS also influences an additional regulatory pathway that likely counteracts the HapR-dependent stimulus. One possibility is that LuxS influences post-transcriptional regulation of RpoS, which is known to be governed by a variety of sRNAs. The dominant phenotype of the cqsA mutation in the cqsA luxS double mutant might then indicate that such regulation is only effective after activation of RpoS transcription (e.g, via HapR). The fact that LuxS is thought to be an agent of interspecies, rather than intraspecies, quorum sensing (Federle and Bassler, 2003), also suggests that the presence of species other than V. cholerae will suppress expression of cluster III chemotaxis genes. Future studies exploring the process by which LuxS negatively regulates cluster III expression should provide a more complete understanding of the complexity of V. cholerae quorum sensing. Such studies might also enable identification of additional stimuli associated with expression of cluster III or provide clues as to the cellular role of cluster III signaling complexes.
Experimental Procedures
Growth conditions and media
V. cholerae, and E. coli were grown in LB media or on LB agar plates at 37°C containing antibiotics in the following concentrations: streptomycin 200 μg/ml; kanamycin 50 μg/ml; ampicillin 100 μg/ml; carbenicillin 50 μg/ml; chloramphenicol 20 μg/ml for E. coli and 5 μg/ml for V. cholerae. For experiments in minimal medium, V. cholerae was grown two different media; MSR6 minimal medium: (50 mM KH2PO4, 50 mM Na2HPO4, 7.5 mM (NH4)2SO4, 2 mM MgSO4, 0.1 mM CaCl2, 25 μM FeSO4) supplemented with varying concentrations of D-glucose and succinate as indicated in the figures and figure legends; M9 minimal medium supplemented with 0.2% glycerol, and 1 μg/mL thiamine.
Strains and plasmids
The strains and plasmids used in this study are listed in Table 1. Primers used are listed in Table 2. E. coli strains DH5αλpir and SM10λpir were used for cloning. E. coli strain SM10λpir was used to transfer DNA into V. cholerae by conjugation (Miller and Mekalanos, 1988). Throughout this study all V. cholerae strains used were derived from the El Tor clinical isolate C6706. Construction of V. cholerae deletion or insertion mutants was performed with standard allele exchange techniques using derivatives of plasmid pCVD442 (Donnenberg and Kaper, 1991). Strain SR9 was created by insertion of cfp-cheW3 on the chromosome replacing the cheW3 at its native locus using plasmid pSR1011 in strain C6706. Strain SR15 was created by insertion of yfp-cheW1 and cfp-cheW3 on the chromosome and replacing the cheW1 and cheW3 at their native locus using plasmids pSR1010 and pSR1011 respectively in strain C6706. Strain SR29 was created by deleting the entire cluster III operon using plasmid pSR1158 in strain C6706. SR36 was created by consecutive deletion of chemotaxis clusters I and II in strain SR9 using plasmids pSR1157 and pSR1020 respectively. Strains SR47, SR48, and SR49 was created by insertion of cfp-cheW3 on the chromosome replacing the cheW3 at its native locus using plasmid pSR1011 in strains MM883, MM893, and KSK1059 respectively. Strain SR71 was created by deletion of rpoS in strain C6706 using plasmid pCVD442-ΔrpoS. Strain SR71 was created by deletion of hapR in strain C6706 using plasmid pSR1217. Strains SR76, SR80, SR81, SR82, and SR83 were created by insertion of rluc8-cheW3 on the chromosome replacing the cheW3 at its native locus using plasmid pSR1213 in strains C6706, MM883, MM893, KSK1059, and SR71 respectively. Plasmid pSR1228 was constructed by PCR amplification of the vca1094 (cheW3) gene using primers CheW3-III-1-cw / CheW3-III-2-ccw and chromosomal DNA from V. cholerae as template. The PCR product was digested with BsrGI and HincII and ligated into the equivalent sites of plasmid pMF391 resulting in plasmid pSR1228. Plasmid pSR1224 was constructed by PCR amplification of the vca1095 (cheA3) gene using primers CheA3-III-cw/CheA3-III-ccw and chromosomal DNA from V. cholerae as template. The PCR product was digested with BsrGI and XbaI and ligated into the equivalent sites of plasmid pMF390 resulting in plasmid pSR1224. Plasmid pSR1011 was constructed by PCR amplification of the up- and down-stream regions of vca1094 (cheW3) using primers CheW3-a/CheW3-bb and CheW3-cc/CheW3-d respectively and V. cholerae chromosomal DNA as template. In a third PCR reaction cfp-vca1094 was amplified using primers CheW3-b/CheW3-c and plasmid pSR1228 as template. A fourth PCR was then performed using primers CheW3-a/CheW3-d and products from all three of the former PCR reactions as template. The resulting PCR product was digested with XbaI and inserted into the equivalent site in pCVD442 resulting in plasmid pSR1011.
Table 1.
Strain and plasmid list.
| Strain name | Genotype | Reference |
|---|---|---|
| Vibrio cholerae C6706 (wild type) | Clinical isolate | |
| SR6 | C6706 lacZ- ΔcheW1::yfp-cheW1 | (Ringgaard et al., 2011) |
| SR9 | C6706 lacZ- ΔcheW3::cfp-cheW3 | This work |
| SR12 | C6706 lacZ- ΔhapR ΔcheW3::cfp-cheW3 | |
| SR15 | C6706 lacZ- ΔcheW1::yfp-cheW1 ΔcheW3::cfp-cheW3 | This work |
| SR28 (Δcluster II) | C6706 lacZ- ΔcheY3 ΔcheZ ΔcheA2 ΔcheB2 ΔparC ΔparP ΔcheW1 | (Hatzios et al., 2012) |
| SR29 (Δcluster III) | C6706 Δvca1088 Δvca1090 Δvca1091 Δvca1092 Δvca1093 Δvca1094 Δvca1095 Δvca1096 | This work |
| SR36 (Δclusters I and II) | C6706 lacZ- ΔcheW3::cfp-cheW3 Δvc1394 Δvc1396 Δvc1397 Δvc1398 Δvc1399 Δvc1400 Δvc1401 Δvc1402 Δvc1403 Δvc1404 Δvc1405 Δvc1406 Δvc2059 Δvc2060 Δvc2061 Δvc2062 Δvc2063 Δvc2064 Δvc2065 | This work |
| SR47 | C6706 ΔcqsA ΔluxS ΔcheW3::cfp-cheW3 | This work |
| SR48 | C6706 ΔcqsA ΔcheW3::cfp-cheW3 | This work |
| SR49 | C6706 ΔluxS ΔcheW3::cfp-cheW3 | This work |
| SR71 | C6706 lacZ- ΔrpoS ΔcheW3::cfp-cheW3 | This work |
| SR76 | C6706 lacZ- ΔcheW3::rluc8-cheW3 | This work |
| SR80 | C6706 ΔcqsA ΔluxS ΔcheW3::rluc8-cheW3 | This work |
| SR81 | C6706 ΔcqsA ΔcheW3::rluc8-cheW3 | This work |
| SR82 | C6706 ΔluxS ΔcheW3::rluc8-cheW3 | This work |
| SR83 | C6706 lacZ- ΔcheW3::cfp-cheW3 ΔrpoS | This work |
| SR84 | C6706 lacZ- ΔcheW3::rluc8-cheW3 ΔrpoS | This work |
| SR89 | C6706 lacZ- ΔcheW3::rluc8-cheW3 ΔhapR | This work |
| MM883 | C6706 ΔcqsA ΔluxS | (Miller et al., 2002) |
| MM893 | C6706 ΔcqsA | (Miller et al., 2002) |
| KSK1059 | C6706 ΔluxS | (Miller et al., 2002) |
| Escherichia coli DH5αλpir | ||
| Escherichia coli SM10λpir |
| Plasmid name | Relevant genotype / description | Reference |
|---|---|---|
| pCVD442 | (Donnenberg and Kaper, 1991) | |
| pCVD442-ArpoS | Plasmid for deletion of rpoS | |
| pSR1010 | Plasmid for insertion of yfp-cheW1 on the chromosome | (Ringgaard et al., 2011) |
| pSR1011 | Plasmid for insertion of cfp-cheW3 on the chromosome | This work |
| pSR1020 | Plasmid for deletion of chemotaxis cluster II | (Hatzios et al., 2012) |
| pSR1033 | PBAD::yfp-cheW1 | (Ringgaard et al., 2011) |
| pSR1157 | Plasmid for deletion of chemotaxis cluster I | (Hatzios et al., 2012) |
| pSR1157 | Plasmid for deletion of chemotaxis cluster III | (Hatzios et al., 2012) |
| pSR1213 | Plasmid for insertion of rluc8-cheW3 on the chromosome | This work |
| pSR1217 | Plasmid for deletion of hapR | This work |
| pSR1228 | PBAD::cfp-vca1094 (cheW3) | This work |
| pSR1224 | PBAD::yfp-vca1095 (cheA3) | This work |
| pMF390 | PBAD::yfp | (Yamaichi et al., 2007) |
| pMF391 | PBAD::cfp | (Yamaichi et al., 2007) |
Table 2.
Primers list.
| Primer name | Primer sequence |
|---|---|
| CheW3-III-1-cw | ccccgagctctgtacaagatgaattcagcgaatttgaccacatc |
| CheW3-III-2-ccw | cccctctagatcatgcttgtccctcattaaggatt |
| CheA3-III-cw | ccccgagctctgtacaagatggctttagatatggaacaactgc |
| CheA3-III-ccw | cccctctagactatgccgctttgcctttggtc |
| CheW3-a | cccctctagaatattcgcatgctaccgatgagtt |
| CheW3-b | caaagcggcataggagcaggtatggtgagcaagggcgagga |
| CheW3-c | cgaaactgagaaattcacgctgcatcatgcttgtccctcattaaggat |
| CheW3-d | cccctctagacatcaaattcgtcgtcatgccttc |
| CheW3-bb | tcctcgcccttgctcaccatacctgctcctatgccgctttg |
| CheW3-cc | atccttaatgagggacaagcatgatgcagcgtgaatttctcagtttcg |
Fluorescence and time-lapse microscopy
Fluorescence microscopy was carried out essentially as described in references (Ringgaard et al., 2009; Ringgaard et al., 2011); cells were mounted on 1% agarose pads in 20% LB on microscope slides. Microscopy was performed using a Zeiss Axioplan 2 microscope equipped with a 100x a-plan lens and Hamamatsu cooled CCD camera. Microscopy of CFP-CheW3 in strain SR29 was performed using a Nikon eclipse Ti inverted microscope equipped with a 100x lens and an Andor Zyla sCMOS cooled camera. Microscopy images were analyzed using ImageJ imaging software and the percentage of cells with polar foci was enumerated by hand using the cell counter plug-in for ImageJ.
To obtain a detailed profile for CFP-CheW3 localization during growth into stationary phase, we diluted 100 μL of an overnight culture (late stationary phase) into 25 mL fresh LB medium and followed growth by measuring optical density. Furthermore, each sample was analyzed by fluorescence microscopy and the percentage of cells with CFP-CheW3 cluster was counted. For microscopy at low optical densities 5 mL culture was harvested and resuspended in 5-50 μL PBS (depending on cell density) and immediately analyzed by fluorescence microscopy.
For the carbon catabolite repression assay, cells were grown in MSR6 medium supplemented with 0.04% w/v D-glucose and 0.4% w/v succinate. Due to catabolite repression, cells will initially only metabolize D-glucose, however, when D-glucose is used up they will adjust and start metabolizing succinate. 100 μL of an overnight culture from MSR6 medium was diluted into 25 mL fresh MSR6 medium. Growth was followed growth by measuring optical density. Furthermore, each sample was analyzed by fluorescence microscopy and the percentage of cells with a CFP-CheW3 cluster was counted.
Infection experiments of infant rabbits by V. cholerae
Infant rabbit infections were carried out as previously described (Ritchie et al., 2010) with minor variation. Briefly, overnight cultures of V. cholerae were diluted 1:100 in LB and shaken at 37°C for 3 hours. The three-hour cultures were harvested by centrifugation and resuspeneded in 2.5% Sodium Bicarbonate (pH 9) at a final concentration of ~2×109 CFU/mL. Rabbits were treated with Cimetidine (50mg/kg, intraperitoneal injection) 3 hours prior to orogastric inoculation with 109 CFU of V. cholerae. Rabbits were monitored and sacrificed immediately following the onset of disease symptoms: diarrhea and staining of the ventral surface. Onset ranged from 14-20 hours. The rabbits that didn't develop symptoms were sacrificed by 24 hours. Following removal of the entire gastrointestinal tract at necropsy, cecal fluid was extracted using needle and syringe, and tissue homogenates were spread on LB agar plates with 200 μg/mL Streptomycin for enumeration of bacterial colonization.
Renilla luciferace assays
For single read stationary phase experiments 5mL LB medium was inoculated with a bacterial colony of the relevant strain. Cultures were incubated at 37°C over night (approximately 16 hours) with shaking. Next day 200 μL cells were sampled and added coelenterazine to a final concentration of 7.5 μM. The 200 μL cells were then added to a well in a 96 well microtiter plate. Samples were then incubated at ambient temperature for 30 minutes to allow for the luciferase generated luminescence signal to stabilize (Hatzios et al., 2012). The total luminescence was then measured for each well with an integration time of 2 seconds using a SpectraMax L Luminescence Microplate Reader from Molecular Devices.
For time course and combined OD600/luminescence experiments, 10 μL cells were added to 1 mL LB with a final concentration of coelenterazine of 7.5 μM. Three times 200 μL cells for each strain were then added to a well in a 96 well microtiter plate. Again, samples were then incubated at ambient temperature for 30 minutes to allow for the luciferase generated luminescence signal to stabilize. Using a Promega GloMax®Multi Detection System with fluorescence and luminescence module the OD600 and the total luminescence measured over time for each well with an integration time of 2 seconds for the luminescence readings.
Western blotting to determine presence of CFP-CheW3
V. cholerae strain SR9 was inoculated in 5 mL LB and incubated overnight at 37°C shaking. 100 μL was then transferred to 50 mL of fresh LB medium at 37°C and shaking. OD600 was measured and samples taken for western blotting as a function of time. The amount of culture harvested and analyzed was normalized for OD600. Samples were then analyzed by SDS-PAGE and western blotting was performed using JL-8 anti-GFP antibodies, which also recognizes CFP. As a loading control samples were also analyzed by western blotting using anti bodies against the flagellar protein FlaA.
RNAseq experiment
RNA-seq was performed basically as described in (Livny et al., 2014). RNA samples were generated from mid-exponential phase (O.D. 600 = ~0.4 – 0.6) and stationary phase (overnight) cultures of C6706 grown in LB. Libraries for each condition were prepared in duplicate from biological replica cultures.
Acknowledgements
We are thanking Dr. Kathrin Schirner for comments on the manuscript and suggestions for experiments. We thank Sören Abel for help and advice regarding rabbit experiments. This work was funded by NIH grant R37 AI-042347 and HHMI (MKW). SR was funded with a postdoctoral fellowship from the Villum Kann Rasmussen foundation. This work was supported by the Max Planck Society (SR).
Footnotes
The authors declare no conflict of interest.
Reference list
- Battesti A, Majdalani N, Gottesman S. The RpoS-Mediated General Stress Response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Bauer CE. A che-like signal transduction cascade involved in controlling flagella biosynthesis in Rhodospirillum centenum. Mol Microbiol. 2005;55:1390–402. doi: 10.1111/j.1365-2958.2005.04489.x. [DOI] [PubMed] [Google Scholar]
- Boin M. a, Austin MJ, Häse CC. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett. 2004;239:1–8. doi: 10.1016/j.femsle.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci U S A. 2004;101:5018–23. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol. 2005;3:611–20. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae Deletion Mutant of Enteropathogenic Escherichia coli by Using a Positive-Selection Suicide Vector. J Bacteriol. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, Bassler BL. Interspecies communication in bacteria. J Clin Invest. 2003;112:1291–1299. doi: 10.1172/JCI20195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink KK, Kobayashi R, Kawagishi I, Häse CC. Analyses of the Roles of the Three cheA Homologs in Chemotaxis of Vibrio cholerae. 2002;184:1767–1771. doi: 10.1128/JB.184.6.1767-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer R, Chen P-Y, Armitage JP, Reinert G, Deane CM. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol. 2010;4:3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzios SK, Ringgaard S, Davis BM, Waldor MK. Studies of Dynamic Protein-Protein Interactions in Bacteria Using Renilla Luciferase Complementation Are Undermined by Nonspecific Enzyme Inhibition. PLoS One. 2012;7:e43175. doi: 10.1371/journal.pone.0043175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen J. a, Nelson WC, Clayton R. a, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–83. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath G, Hyakutake A, Yamamoto K, Ebisawa T, Nakamura T, Nishiyama S-I, et al. Hypoxia-induced localization of chemotaxis-related signaling proteins in Vibrio cholerae. Mol Microbiol. 2014 doi: 10.1111/mmi.12887. doi: 10.1111/mmi.12887. [DOI] [PubMed] [Google Scholar]
- Hyakutake A, Homma M, Austin MJ, Boin MA, Häse CC, Kawagishi I. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J Bacteriol. 2005;187:8403–8410. doi: 10.1128/JB.187.24.8403-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joelsson A, Kan B, Zhu J. Quorum sensing enhances the stress response in Vibrio cholerae. Appl Environ Microbiol. 2007;73:3742–6. doi: 10.1128/AEM.02804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Zhou X, Mandlik A, Hubbard T, Davis BM, Waldor MK. Comparative RNA-Seq based dissection of the regulatory networks and environmental stimuli underlying Vibrio parahaemolyticus gene expression during infection. Nucleic Acids Res. 2014;42:12212–12223. doi: 10.1093/nar/gku891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar Localization of the Chemotreceptor Complex in Escherichia coli. Cell. Science (80-) 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Clark D. Brock biology of microorganisms. (9th ed.) 2000 [Google Scholar]
- Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10:165–74. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014;11:1–14. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Skorupski K, Lenz D. Parallel Quorum Sensing Systems Converge to Regulate Virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet Y. a, Alvarez D, Ringgaard S, Andrian U.H. von, Davis BM, Waldor MK. Insights into Vibrio cholerae Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria. PLoS Pathog. 2014;10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AT, Dolganov N. a, Otto G, Miller MC, Wu CY, Schoolnik GK. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, Schirner K, Davis BM, Waldor MK. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 2011;25:1544–1555. doi: 10.1101/gad.2061811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, Zepeda-Rivera M, Wu X, Schirner K, Davis BM, Waldor MK. ParP prevents dissociation of CheA from chemotactic signaling arrays and tethers them to a polar anchor. Proc Natl Acad Sci U S A. 2014;111:E255–64. doi: 10.1073/pnas.1315722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, Zon J. van, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci U S A. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie J, Rui H, Bronson R, Waldor M. Back to the future: studying cholera pathogenesis using infant rabbits. MBio. 2010;1:e00047–10. doi: 10.1128/mBio.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Bassler B. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–51. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in Chemotaxis Mechanisms among the Bacteria and Archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–37. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Yamaichi Y, Fogel M. a, Waldor MK. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:630–635. doi: 10.1073/pnas.0608341104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–56. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–72. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]