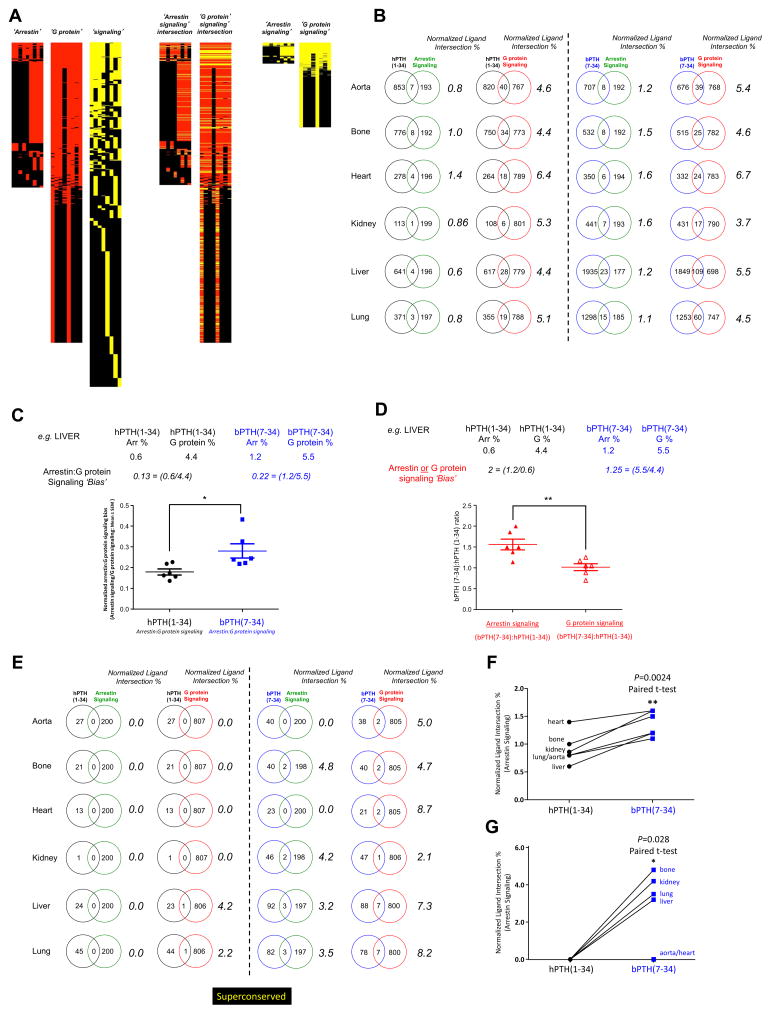
Fig. 5.
bPTH(7-34) in vivo transcriptomic activity is strongly associated with arrestin-biased signaling. (A) Generation of arrestin, G protein and cellular signaling theoretical datasets using latent semantic indexing. A PubMed-based database of >2×106 indexed abstracts was used to generate lists of genes, from the whole murine genomic background set associated with the input interrogation terms, semantically-linked to ‘arrestin’, ‘G protein’ or ‘cell signaling’-related text terms. The intersections between the ‘arrestin’ or ‘G protein’ datasets with the ‘cellular signaling’ dataset were used to create the resultant theoretical ‘arrestin signaling’ and ‘G protein signaling’ datasets. (B) Percentage intersection scoring (normalized for different dataset sizes) for both hPTH(1-34) and bPTH(7-34) transcriptomic response datasets (full significantly-regulated datasets) with either the ‘arrestin signaling’ or ‘G protein signaling’ theoretical datasets. (C) Arrestin:G protein signaling bias analysis for hPTH(1-34) and bPTH(7-34). An example calculation from the intersection analysis results for liver data is depicted. (D) Arrestin or G protein signaling bias analysis for hPTH(1-34) and bPTH(7-34). An example calculation from the intersection analysis results for liver is depicted. (E) Percentage intersection scoring (normalized for different dataset sizes) for both hPTH(1-34) and bPTH(7-34) superconserved transcriptomic response datasets with either the ‘arrestin signaling’ or ‘G protein signaling’ theoretical datasets. (F) Percentage intersection analysis (paired t-test across tissues) for the whole significant transcriptome data revealed a significantly greater (p=0.0024) degree of intersection between bPTH(7-34) empirical data with the ‘arrestin signaling’ theoretical dataset compared to hPTH(1-34) data. (G) Percentage intersection analysis (paired t-test across tissues) for the superconserved transcriptome data again revealed a significantly greater (p=0.028) degree of intersection between bPTH(7-34) empirical data with the ‘arrestin signaling’ theoretical dataset compared to hPTH(1-34) data.