Fly Dicer-2 exhibits a non–RNA interference function in the posttranscriptional modulation of Toll protein expression and Toll signaling.
Keywords: Dicer-2, invertebrate immunity, RNA silencing, Toll signaling, posttranscriptional gene regulation
Abstract
Dicer-2 is the central player for small interfering RNA biogenesis in the Drosophila RNA interference (RNAi) pathway. Intriguingly, we found that Dicer-2 has an unconventional RNAi-independent function that positively modulates Toll immune signaling, which defends against Gram-positive bacteria, fungi, and some viruses, in both cells and adult flies. The loss of Dicer-2 expression makes fruit flies more susceptible to fungal infection. We further revealed that Dicer-2 posttranscriptionally modulates Toll signaling because Dicer-2 is required for the proper expression of Toll protein but not for Toll protein stability or Toll mRNA transcription. Moreover, Dicer-2 directly binds to the 3′ untranslated region (3′UTR) of Toll mRNA via its PAZ (Piwi/Argonaute/Zwille) domain and is required for protein translation mediated by Toll 3′UTR. The loss of Toll 3′UTR binding activity makes Dicer-2 incapable of promoting Toll signaling. These data indicate that the interaction between Dicer-2 and Toll mRNA plays a pivotal role in Toll immune signaling. In addition, we found that Dicer-2 is also required for the Toll signaling induced by two different RNA viruses in Drosophila cells. Consequently, our findings uncover a novel RNAi-independent function of Dicer-2 in the posttranscriptional regulation of Toll protein expression and signaling, indicate an unexpected intersection of the RNAi pathway and the Toll pathway, and provide new insights into Toll immune signaling, Drosophila Dicer-2, and probably Dicer and Dicer-related proteins in other organisms.
INTRODUCTION
Innate immunity plays critical roles in the detection and eradication of a wide range of microbial pathogens such as bacteria, fungi, and viruses, and serves as the first line of defense against microbial invasion (1–4). The fruit fly Drosophila melanogaster, like other insects, lacks acquired immune responses and completely relies on innate immunity to defend against pathogens (4). The innate immune defenses of Drosophila include three critical pathways: Toll, immune deficiency (IMD), and RNA interference (RNAi).
The Toll and IMD pathways regulate the activation of the nuclear factor κB (NF-κB) transcription factors DIF/Dorsal and Relish, respectively (5–7). The Toll pathway is induced by fungi or Gram-positive bacteria, leading to the activation of the Toll transmembrane receptor through a cascade of extracellular proteolytic events. The activated Toll finally activates DIF/Dorsal through a signaling cascade via dMyD88, Pelle, Tube, and Cactus, resulting in the transcriptional induction of multiple antimicrobial peptide (AMP) genes, including Drosomycin (Dros) (5, 8, 9). The IMD pathway is activated by Gram-negative bacteria similarly to Toll signaling, leading to Relish activation and transcriptional induction of some AMPs, including Diptericin (Dipt) (8, 10). On the other hand, the RNAi pathway is a potent antiviral defense in which the double-stranded RNA (dsRNA) region of viral replicative intermediates is detected and processed by dsRNA-specific endoribonuclease Dicer into 21- to 23-nucleotide small interfering RNAs (siRNAs). The virus-derived siRNAs are transferred by the Dicer-R2D2 complex to Argonaute (AGO) protein in the RNA-induced silencing complex (RISC), which guides the specific pairing and destruction of homologous viral RNA in infected cells (11–16). All three innate immune pathways are conserved in insects and mammals. The Drosophila IMD pathway is homologous to the mammalian tumor necrosis factor pathway; the Toll pathway is similar to the mammalian Toll-like receptor (TLR)–interleukin-1 pathway; and the RNAi pathway has been shown to be an antiviral mechanism in mammals (17, 18).
Dicer-2 (Dcr-2) is the sole siRNA-producing Dicer protein of Drosophila that plays essential roles as a pattern recognition receptor (PRR) and as a component of RISC in antiviral RNAi defense in Drosophila (14, 19). Like most metazoan Dicer proteins, Drosophila Dcr-2 is composed of an N-terminal DExD/H-box RNA helicase domain, a DUF283 domain, a PAZ (Piwi/Argonaute/Zwille) domain, two tandem RNase III domains, and a C-terminal dsRNA binding domain (dsRBD). A previous study reported that Dcr-2 can mediate the induction of the antiviral protein Vago, which controls Drosophila C virus (DCV) infection, independently of RNAi; however, Dcr-2–mediated Vago induction also occurs independently of either the Toll pathway or the IMD pathway in Drosophila (20). Moreover, a previous sequence analysis of Dcr-2 by Deddouche et al. (20) revealed that Dcr-2 is phylogenically related to mammalian retinoic acid–inducible gene I (RIG-I). Besides inducing type I interferon (a canonical function) (21), RIG-I also posttranscriptionally regulates NF-κB expression (22). Thus, it is interesting to examine whether Dcr-2 is also involved in some non-RNAi immune pathways in Drosophila.
Here, we report that Dcr-2 is also required for Toll immune signaling independently of RNAi both in cells and in adult flies. The loss of Dcr-2 expression renders fruit flies more susceptible to fungal infection. We further revealed that Dcr-2 positively modulates Toll signaling at the posttranscriptional level because Dcr-2 is required for the proper expression of Toll protein but not for Toll protein stability or Toll mRNA transcription. Moreover, Dcr-2 directly binds to the 3′ untranslated region (3′UTR) of Toll mRNA via its PAZ domain and is required for protein expression mediated by Toll 3′UTR. The loss of Toll 3′UTR binding activity makes Dcr-2 incapable of promoting Toll signaling. These results indicate that the interaction between Dcr-2 and Toll mRNA plays a pivotal role in the posttranscriptional modulation of Toll protein expression and Toll signaling. Furthermore, we uncovered that Dcr-2 is required for the Toll signaling induced by two different RNA viruses, Flock House virus (FHV) and vesicular stomatitis virus (VSV), in cultured Drosophila cells. Consequently, our findings uncover an unconventional RNAi-independent function of Dcr-2 in the posttranscriptional regulation of Toll protein expression and indicate an unexpected intersection of the RNAi pathway and the Toll pathway. These findings also suggest that Dicer and Dicer-related proteins in Drosophila and other organisms, including plants and mammals, play additional roles in innate immunity and other processes.
RESULTS
Dcr-2 is required for Toll innate immune signaling in Drosophila S2 cells
Toll immune signaling can be activated by infection with fungi, Gram-positive bacteria, or certain viruses (23–26). All these microbial pathogens could significantly induce the expression of Dros (23, 24), which is a downstream AMP specific for the Toll pathway.
To determine whether Dcr-2 is involved in Toll immune signaling, we used two dsRNAs (numbers 1 and 2) targeting nonoverlapping regions of the dcr-2 gene to knock down endogenous Dcr-2 in Drosophila S2 cells. Moreover, we also designed egfp dsRNA and two ago2 dsRNAs (numbers 1 and 2) as controls. The activity of Toll immune signaling was monitored by measuring the transcription level of Dros. Our results showed that Dcr-2 knockdown decreased basal Dros transcription in cultured S2 cells [Fig. 1, A (lane 3) and B (lane 3)], whereas AGO2 or enhanced green fluorescent protein (EGFP) knockdown did not decrease the level of Dros mRNA [Fig. 1, A (lanes 2 and 4) and B (lanes 2 and 4)].
Fig. 1. Dcr-2 is required for Toll signaling in Drosophila S2 cells.
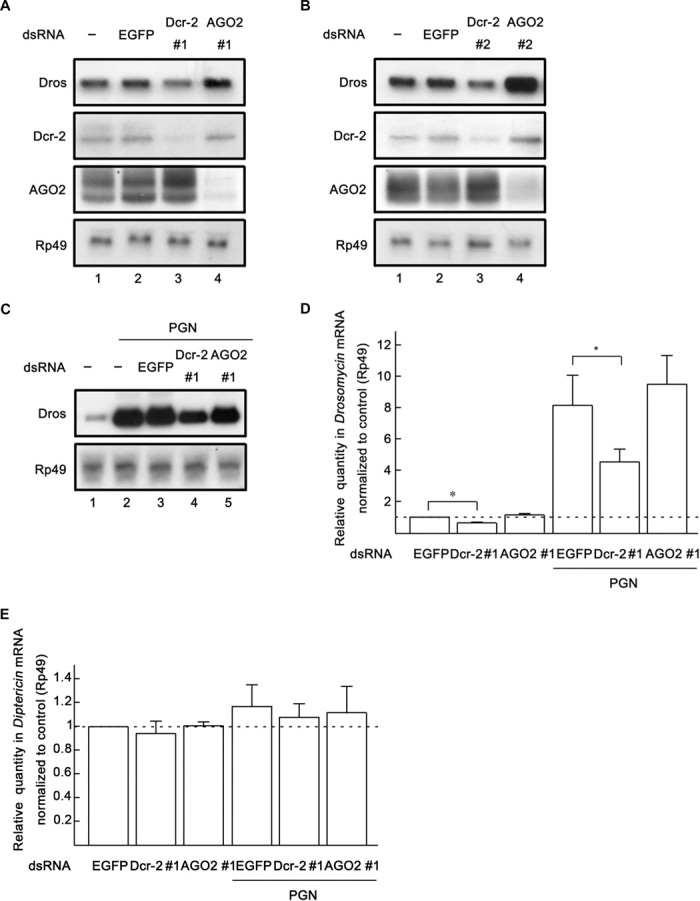
(A and B) Cultured S2 cells were transfected with dsRNAs against the indicated genes. Total RNA extracts were prepared from the cells and detected via Northern blots using the indicated probes. (C) Cultured S2 cells were transfected with dsRNAs against the indicated genes and treated with Lys-PGN (100 μg/ml; lanes 2 to 5) or water (lane 1) for 20 hours. Total RNA extracts were prepared from the cells and detected via Northern blots using the indicated probes. (D and E) Cultured S2 cells were transfected with dsRNAs and treated with Lys-PGN as indicated. Total RNA extracts were prepared for qRT-PCR assay of Dros (D) or Dipt (E) mRNA (normalized to Rp49; n = 3; *P < 0.05, two-tailed Student’s t test; error bars, SD).
Previous studies demonstrated that Toll signaling in cultured S2 cells can be activated by lysine-type peptidoglycan (Lys-PGN) (27), which is an important component of Gram-positive bacterial envelopes. After we determined that Dcr-2 is involved in the induction of Dros transcription in S2 cells, we sought to determine the effects of Dcr-2 under microbial infection. To this end, we treated S2 cells with Lys-PGN (extract from Staphylococcus aureus) to simulate Gram-positive bacterial infection and assessed the effects of Dcr-2 knockdown on peptidoglycan (PGN)–induced Toll signaling. As expected, PGN treatment dramatically induced Dros transcription (Fig. 1C, lanes 1 and 2). Dcr-2 knockdown decreased PGN-induced Toll signaling (that is, Dros transcription) in cultured S2 cells (Fig. 1C, lane 4), whereas AGO2 or EGFP knockdown had no such effect (Fig. 1C, lanes 3 and 5).
We further examined the role of Dcr-2 in Dros induction in S2 cells using quantitative reverse transcription polymerase chain reaction (qRT-PCR). Consistent with the results obtained with Northern blots of Dros mRNA, our qRT-PCR data showed that dcr-2 dsRNA significantly reduced the induction of Dros in both nontreated and PGN-treated S2 cells, whereas dsRNA against AGO2 did not reduce Dros induction (Fig. 1D). On the other hand, neither dcr-2 knockdown nor ago2 knockdown by dsRNA affected the induction of Dipt (the AMP specific for the IMD pathway) (Fig. 1E) (28), indicating that Dcr-2 is not involved in the IMD pathway. Moreover, previous studies showed that the transient overexpression of certain Toll pathway components, such as dMyD88 or Pelle alone, can induce Dros transcription (29). To further confirm the role of Dcr-2 in Toll signaling, we overexpressed Flag-tagged EGFP (EGFP-Flag), Flag-tagged Dcr-2 (Dcr-2–Flag), and Flag-tagged AGO2 (AGO2-Flag) in cultured S2 cells. Our results showed that overexpression of Dcr-2 resulted in the induction of Dros, as measured by Northern blots (fig. S1A) and qRT-PCR (fig. S1B), whereas overexpression of EGFP or AGO2 did not result in Dros induction (fig. S1A). Altogether, our data show that Dcr-2 is involved in the Toll pathway, but not in the IMD pathway, in cultured S2 cells.
Dcr-2 is required for Toll signaling and resistance to microbial infection in Drosophila
To assess the role of Dcr-2 in vivo, we sought to determine whether Dcr-2 is also required for Toll signaling in adult Drosophila flies. To this end, we used transgenic flies carrying a dcr-2 dsRNA construct driven by the Gal4/UAS promoter. Our results showed that ubiquitous expression of Gal4 with the Actin driver in UAS-dsRNA (Dcr-2) flies resulted in successful knockdown of Dcr-2 (Fig. 2A, middle) and dramatically inhibited the induction of Dros transcription (Fig. 2A, lanes 1 and 2). Adult flies were subjected to a microbial challenge test in which they were pricked with a Gram-positive bacterium, Micrococcus luteus. Consistent with previous studies (14, 29), M. luteus significantly induced Dros transcription (Fig. 2A, lanes 1 and 3). In the case of the M. luteus challenge, the ubiquitous knockdown of Dcr-2 in flies also dramatically inhibited the induction of Dros transcription (Fig. 2A, lanes 3 and 4). Similar results were also obtained via qRT-PCR assay (Fig. 2B).
Fig. 2. Dcr-2 is required for Toll activation in adult flies.
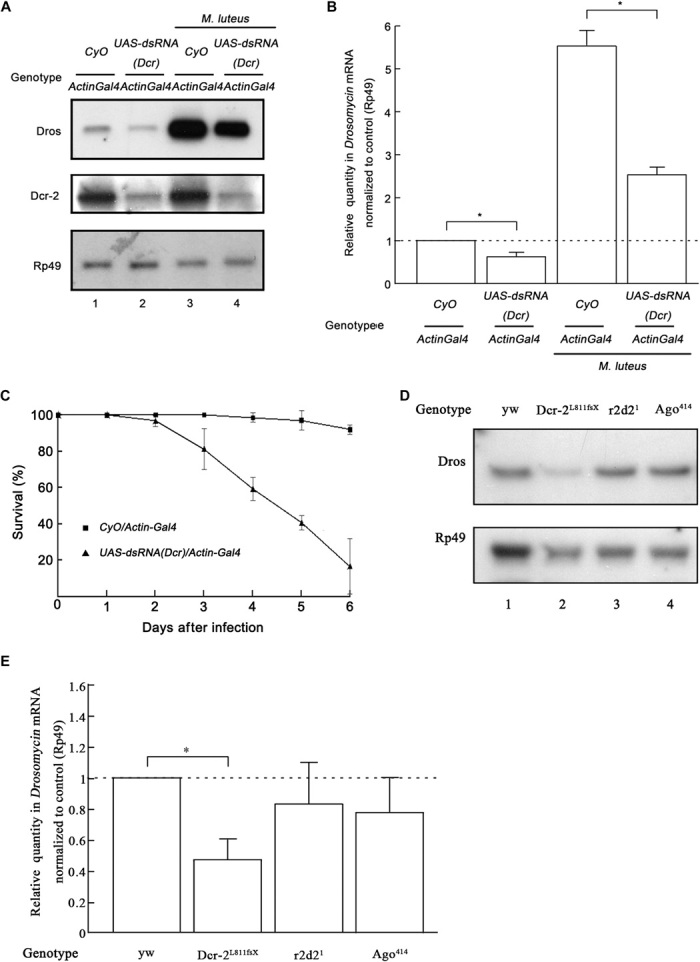
(A and B) Six hours after the M. luteus challenge, total RNA extracts were prepared from adult flies with the indicated genotypes and treatment and subjected to Northern blots using the indicated probes (A) or to qRT-PCR of Dros mRNA (normalized to Rp49; n = 3; *P < 0.05, two-tailed Student’s t test; error bars, SD; each group contains 10 female flies and 10 male flies) (B). CyO, second chromosome balancer with curly wings marker. (C) Survival of adult flies with the indicated genotypes after A. fumigatus infection (n = 3; each group contains 10 female flies and 10 male flies; error bars, SD). Flies that died within 3 hours of infection were not considered in the analysis. (D and E) Total RNA extracts were prepared from adult flies with the indicated genotypes and subjected to Northern blots using the indicated probes (D) or to qRT-PCR of Dros mRNA (normalized to Rp49; n = 3; *P < 0.05, two-tailed Student’s t test; error bars, SD) (E).
Considering that downstream AMPs of Toll immune signaling are required for adult flies’ resistance to infection by Gram-positive bacteria or fungi, we analyzed the survival rate of dcr-2 knockdown transgenic flies infected with Aspergillus fumigatus. Because A. fumigatus is weakly pathogenic to wild-type flies but is harmful to flies with impaired Toll immune signaling, A. fumigatus infection was used by Lemaitre et al. (28) to determine the resistance of different mutant flies to microbial infection. As shown in Fig. 2C, 6 days after A. fumigatus infection, about 80% of dcr-2 knockdown transgenic flies had died whereas flies without dcr-2 knockdown were not markedly affected. These data indicate that the loss of Dcr-2 expression dramatically made adult flies more susceptible to A. fumigatus infection.
Moreover, we used Dcr-2–null allele flies to further confirm the role of Dcr-2 in Toll signaling and used R2D2- and AGO2-null allele flies as controls. Our data showed that the induction of Dros transcription in Dcr-2L811fsX (Dcr-2–null) flies [versus that in yw (yellow white) flies] was significantly inhibited, as determined by Northern blots (Fig. 2D) and qRT-PCR (Fig. 2E), whereas the loss of R2D2 or AGO2 in flies (r2d21 or Ago2414) had no such effect (Fig. 2, D and E). Consistent with data obtained from cultured Drosophila S2 cells, we conclude that Dcr-2, but not the siRNA pathway, is required for the activation of Toll immune signaling.
Dcr-2 is involved in the expression of Toll protein at the posttranscriptional level
After we have determined that Dcr-2 is required for Toll immune signaling in both cultured S2 cells and adult flies, we attempted to determine at which step Dcr-2 affects Toll signaling. To this end, Flag-tagged Toll transgene (Flag-Toll) was ectopically expressed in cultured S2 cells. We observed that the ectopic expression of Flag-Toll successfully rescued the down-regulation of Dros transcription in cultured S2 cells transfected with dcr-2 dsRNA, as determined by Northern blots (Fig. 3A) and qRT-PCR (Fig. 3B). These results indicate that Dcr-2 could affect the Toll signaling pathway upstream of Toll. We next assessed the impact of Dcr-2 knockdown on Toll mRNA transcription. The results of Northern blot analyses showed that the knockdown of Dcr-2 by dsRNA had little effect on the levels of Toll mRNA (Fig. 3C). Similar results were also obtained via qRT-PCR assay (Fig. 3D).
Fig. 3. Dcr-2 is involved in the expression of Toll protein at the posttranscriptional level.
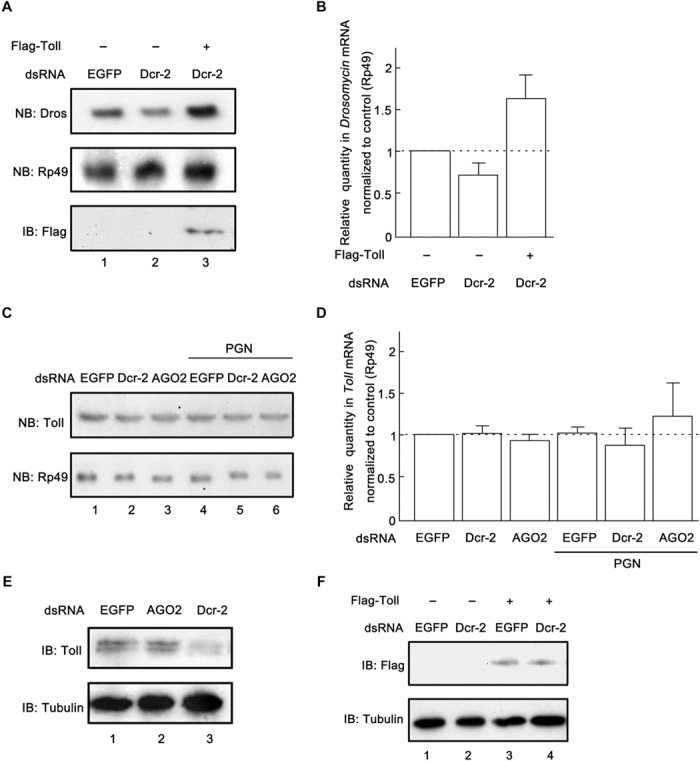
(A) Cultured S2 cells were transfected with empty vector (lanes 1 and 2), the plasmid expressing Flag-Toll (lane 3), and dsRNAs against the indicated genes. Total RNA extracts were prepared and detected via Northern blots using the indicated probes. Cell lysates were prepared and subjected to Western blots using the indicated antibody. (B) RNA extracts were prepared as in (A) for qRT-PCR assay of Dros mRNA (normalized to Rp49; n = 3; error bars, SD). (C and D) Cultured S2 cells were transfected with the indicated dsRNAs and treated with Lys-PGN (100 μg/ml) (C; lanes 4 to 6) or water (C; lanes 1 to 3) for 20 hours, as indicated. Total RNA extracts were prepared for Northern blots using the indicated probes (C) or for qRT-PCR of Toll mRNA (normalized to Rp49; n = 3; error bars, SD) (D). (E) Cultured S2 cells were transfected with dsRNAs against the indicated genes. Cell lysates were prepared and subjected to Western blots using the indicated antibodies. (F) Cultured S2 cells transfected with empty vector (lanes 1 and 2), the plasmid expressing Flag-Toll (lanes 3 and 4), and dsRNAs against the indicated genes. Cell lysates were prepared and subjected to Western blots using the indicated antibodies. NB, Northern blots; IB, immunoblots or Western blots.
Because Dcr-2 knockdown had no impact on the mRNA level of Toll, we sought to determine whether Dcr-2 knockdown affected the protein expression of Toll. Our results showed that Dcr-2 knockdown resulted in an obvious reduction of the protein level of Toll in S2 cells (Fig. 3E). We also found that the exogenous expression of Flag-Toll was not affected by Dcr-2 knockdown (Fig. 3F), indicating that Dcr-2 had no effect on the stability of Toll protein.
Taken together, our data show that Dcr-2 modulates Toll protein expression at the posttranscriptional level but not at the level of Toll mRNA transcription or Toll protein stability.
Dcr-2 directly interacts with the 3′UTR of Toll mRNA
A recent study showed that RIG-I binds to the 3′UTRs of several cellular mRNAs and regulates translation (22). These studies prompted us to speculate that Dcr-2 may also bind to Toll mRNA and further modulate Toll protein expression. To explore this possibility, we performed RNA immunoprecipitation (RNA-IP) using an anti-Flag antibody from the total lysates of Drosophila S2 cells ectopically expressing Dcr-2–Flag (Fig. 4A, lanes 3 and 4). The lysates of cells transfected with empty vector were used as controls (Fig. 4A, lanes 1 and 2). Our results clearly showed that Dcr-2–Flag could bind to Toll mRNA (Fig. 4A, panels 2 and 3, lane 3). Moreover, when the in vitro–transcribed 3′UTR of Toll mRNA was transfected into the cells, the binding of Dcr-2 to Toll mRNA was dramatically inhibited (Fig. 4A, panels 2 and 3, lanes 3 and 4), and Dcr-2 was found to bind to the exogenous Toll 3′UTR (Fig. 4A, top, lane 4). The Northern blot results were further confirmed by qRT-PCR assay (Fig. 4B). To further confirm the identity of the Toll mRNA that was precipitated with Dcr-2–Flag via RNA-IP, we performed RT-PCR to reversely transcribe RNAs precipitated by Dcr-2–Flag using Toll-specific primers. Our results showed that DNA fragments amplified by RT-PCR were only observed when Dcr-2–Flag was ectopically expressed (Fig. 4C, lanes 3 and 4). The presence of exogenous Toll 3′UTR consistently decreased the amount of RT-PCR products (Fig. 4C, lane 4). To exclude the possibility of the Flag tag nonspecifically binding to Toll mRNA, we used EGFP-Flag as a control in the RNA-IP assay. In addition, we also examined whether AGO2-Flag could interact with Toll mRNA. Our results showed that neither EGFP-Flag nor AGO2-Flag was able to bind Toll mRNA (fig. S2A). Moreover, because exogenous Toll 3′UTR can compete with Toll mRNA for Dcr-2 interaction (Fig. 4A), we assessed the impact of Toll 3′UTR on Toll immune signaling. Our results showed that, in the presence of exogenous Toll 3′UTR, the induction of Dros transcription in cultured S2 cells was significantly inhibited (fig. S2, B and C).
Fig. 4. Dcr-2 binds to the 3′UTR of Toll mRNA.
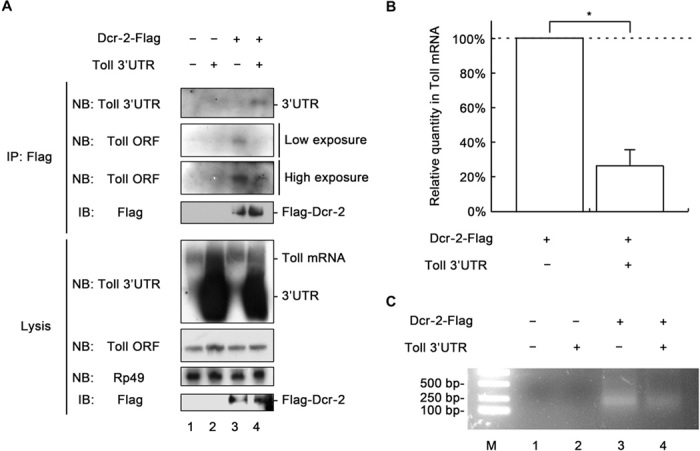
(A) Cultured S2 cells were transfected with empty vector (lanes 1 and 2), the plasmid expressing Dcr-2–Flag (lanes 3 and 4), and in vitro–transcribed Toll 3′UTR (lanes 2 and 4), as indicated. Crude cell lysates were prepared and subjected to RNA-IP using anti-Flag antibody. RNA extracts prepared from precipitates and total cell lysates were subjected to Northern blots, as indicated. Precipitates and total cell lysates were also subjected to Western blots using the indicated antibodies. (B) RNA extracts were prepared from RNA-IP precipitates as in (A) and subjected to qRT-PCR of Toll mRNA (n = 3; *P < 0.01, two-tailed Student’s t test; error bars, SD). (C) RNA extracts were prepared as described in (B) and subjected to RT-PCR with Toll-specific primers. RT-PCR products were analyzed via agarose gel electrophoresis. NB, Northern blots; IB, immunoblots or Western blots.
To further confirm the specificity of the Dcr-2–Toll mRNA interaction, we examined whether Dcr-2 can interact with the mRNAs of some other important factors in the Toll pathway (for example, dMyD88, DIF, and Dorsal) and IMD pathway (for example, PGRP-LC, IMD, and Relish). To this end, the RNA-IP assay was performed using an anti-Flag antibody from cell lysates expressing Dcr-2–Flag, and RNA extracts prepared from RNA-IP precipitates were examined using qRT-PCR. Our data showed that Dcr-2 could bind only to Toll mRNA but not to dMyD88, DIF, Dorsal, PGRP-LC, IMD, or Relish mRNA (fig. S2D).
After we determined that Dcr-2 can bind to the 3′UTR of Toll mRNA, we further examined whether this protein-RNA interaction is direct or indirect. To this end, we expressed and purified 8×His-tagged full-length Dcr-2 in a baculovirus expression system (Fig. 5A), incubated it with an in vitro–transcribed digoxigenin (DIG)–labeled Toll 3′UTR, and subjected it to a gel shift assay. Our data showed that Toll 3′UTR underwent an apparent gel mobility shift when it was incubated with His8–Dcr-2 (Fig. 5B), indicating that Dcr-2 binds to Toll 3′UTR via direct interaction.
Fig. 5. Dcr-2 directly binds to Toll 3′UTR.
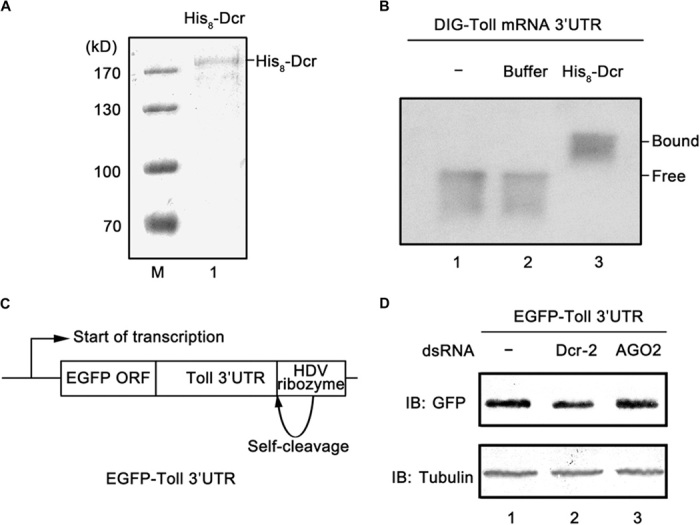
(A) SDS-PAGE analysis of purified His8–Dcr-2 and Coomassie brilliant blue staining. M, molecular mass marker; lane 1, His8–Dcr-2. (B) Gel mobility shift assay was performed to evaluate the capacity of His8–Dcr-2 to bind in vitro–transcribed Toll 3′UTR. Lane 1, Toll 3′UTR alone; lane 2, Toll 3′UTR in reaction buffer; lane 3, Toll 3′UTR in reaction buffer supplemented with His8–Dcr-2. Protein-bound and free RNA are indicated. (C) Schematic diagram of the plasmid that transcribes the mRNA containing the egfp ORF followed by the Toll 3′UTR. (D) The plasmid diagrammed in (C) was cotransfected with dcr-2 (lane 2) or ago2 (lane 3) dsRNA into cultured S2 cells. Cell lysates were prepared and subjected to Western blots using the indicated antibodies.
Furthermore, to assess the role of Toll 3′UTR in Dcr-2–mediated protein expression, we designed a plasmid that can transcribe an mRNA containing an egfp open reading frame (ORF) followed by Toll 3′UTR (Fig. 5C). The plasmid was cotransfected with dcr-2 or ago2 dsRNA into cultured S2 cells for 3 days, and the protein expression of EGFP was examined using Western blots. We found that Dcr-2 knockdown reduced the protein expression of EGFP (Fig. 5D), whereas the same knockdown had no effect on protein expression by an ordinary EGFP expression vector containing a simian virus 40 polyadenylation signal. These data showed that Toll 3′UTR is required for Dcr-2–mediated protein expression.
Altogether, we conclude that Dcr-2 can directly interact with the 3′UTR of Toll mRNA and that this direct interaction between Dcr-2 and Toll mRNA plays a critical role in the posttranscriptional modulation of Toll protein expression.
Dcr-2 directly interacts with Toll 3′UTR via its PAZ domain
Dicer proteins, including Drosophila Dcr-2, contain multiple domains that play distinct roles in Dicer activities (13, 30). The recombinant PAZ domain (Fig. 6A, lane 1), RNase III domain (Fig. 6A, lane 2), dsRBD domain (Fig. 6A, lane 3), and DExD/H-box helicase domain (Fig. 6C, lane 1) of Dcr-2 were expressed and purified to further determine which domain(s) mediates the direct interaction between Dcr-2 and Toll 3′UTR. Each Dcr-2 domain was incubated with in vitro–transcribed DIG-labeled Toll 3′UTR and subjected to a gel shift assay. Our results showed that the recombinant PAZ domain caused a clear gel mobility shift (Fig. 6B, lane 3), whereas the dsRBD and DExD/H-box helicase domains failed to show any interaction with Toll 3′UTR [Fig. 6, B (lane 5) and D (lane 3)]. The presence of the myelin basic protein (MBP) fusion RNase III domain resulted in a dramatic reduction in the level of Toll 3′UTR (Fig. 6B, lane 4), which could have been caused by RNA degradation via its RNase activity. Moreover, the recombinant RNase III domain caused a barely detectable gel mobility shift that was probably attributable to its intrinsic RNA binding activity; however, the direct interaction between Dcr-2 and Toll 3′UTR was unlikely to be mediated by the RNase III domain in an integral Dcr-2 protein because Dcr-2 did not down-regulate either Toll mRNA or Toll signaling, as previously shown (Figs. 1 to 5). To further confirm the specificity of the direct interaction between the PAZ domain and Toll 3′UTR, we performed a gel shift assay that examined whether the recombinant PAZ domain could bind to the in vitro–transcribed 3′UTRs of DIF and Dorsal (the NF-κB transcription factors in the Toll pathway). Our data showed that the PAZ domain had no interaction with the in vitro–transcribed Dif or dorsal 3′UTR (fig. S3, A and B).
Fig. 6. Dcr-2 directly interacts with Toll 3′UTR via its PAZ domain in vitro.
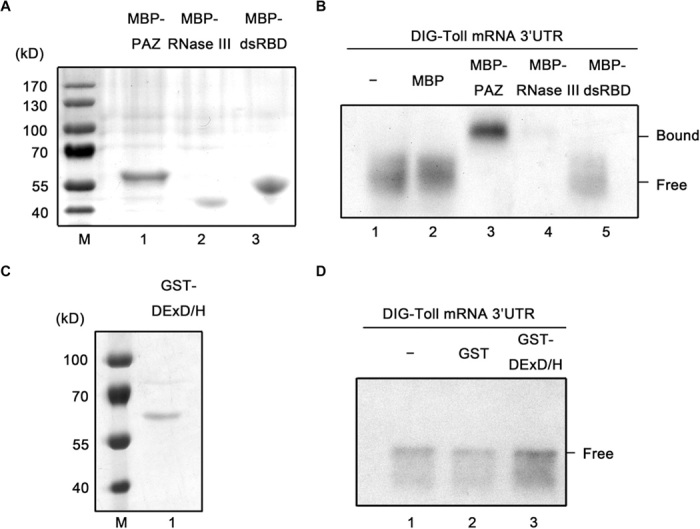
(A) SDS-PAGE analyses of purified MBP-PAZ (lane 1), MBP–RNase III (lane 2), and MBP-dsRBD (lane 3), and Coomassie brilliant blue staining. (B) Gel shift assays were performed to evaluate the capacity of PAZ, RNase III, or dsRBD domain to bind in vitro–transcribed Toll 3′UTR, as indicated. (C) SDS-PAGE analyses of purified GST DExD/H-box helicase domain (lane 1) and Coomassie brilliant blue staining. (D) Gel shift assays were performed to evaluate the capacity of DExD/H-box helicase domain to bind in vitro–transcribed Toll 3′UTR, as indicated. (A and C) M, molecular mass marker.
To determine whether the PAZ domain could interact with Toll mRNA in cultured S2 cells, we performed an RNA-IP assay using an anti-Flag antibody from the total lysates of S2 cells ectopically expressing a Flag-tagged PAZ domain (Fig. 7A, lane 2) or a dsRBD domain (Fig. 7A, lane 3). As shown in Fig. 7A (top), only the Flag-tagged PAZ domain could bind to Toll mRNA. This result was further confirmed by RT-PCR assay (Fig. 7B). Overexpression of the PAZ domain resulted in a dramatic reduction of Dros transcription (Fig. 7A, panel 3). Moreover, cultured S2 cells were transfected with the plasmid, which transcribes an mRNA containing an egfp ORF followed by Toll 3′UTR (Fig. 5C), and with the expression vector for the Flag-tagged PAZ domain. Our results showed that ectopic expression of the PAZ domain dramatically reduced the expression of EGFP (Fig. 7C), indicating that the exogenous PAZ domain can compete with Dcr-2 for Toll 3′UTR, resulting in the reduction of Toll 3′UTR–mediated protein expression. A Dcr-2 mutant with PAZ domain deletion (Dcr-2ΔPAZ-Flag) was used in RNA-IP to further confirm the role of the PAZ domain in Dcr-2 binding to Toll mRNA. Our data showed that deletion of the PAZ domain resulted in the loss of Dcr-2 binding to Toll mRNA (Fig. 7D). Moreover, overexpression of Dcr-2ΔPAZ had no effect on Dros induction (Fig. 7E), indicating that the loss of Toll 3′UTR binding activity makes Dcr-2 incapable of modulating Toll signaling.
Fig. 7. Dcr-2 interacts with Toll mRNA via its PAZ domain in S2 cells.
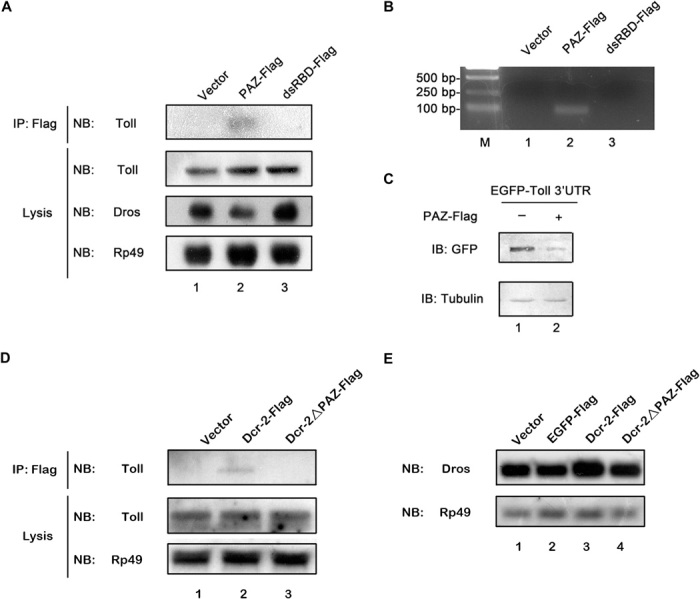
(A) Cultured S2 cells were transfected with empty vector (lane 1), the plasmid expressing Flag-tagged PAZ domain (lane 2), or the Flag-tagged dsRBD domain (lane 3), as indicated. Crude cell lysates were prepared and subjected to RNA-IP using anti-Flag antibody. RNA extracts prepared from precipitates and total cell lysates were subjected to Northern blots, as indicated. (B) RNA extracts from the precipitates in (A) were subjected to RT-PCR with Toll-specific primers. RT-PCR products were analyzed via agarose gel electrophoresis. M, molecular mass marker. (C) The plasmid transcribing mRNA containing the egfp ORF followed by the Toll 3′UTR, as described in Fig. 5C, was cotransfected with empty vector (lane 1) or the plasmid expressing Flag-tagged PAZ domain (lane 2) into cultured S2 cells. Cell lysates were prepared and subjected to Western blots using the indicated antibodies. (D) Cultured S2 cells were transfected with empty vector (lane 1), the plasmid expressing Dcr-2–Flag (lane 2), or Dcr-2ΔPAZ-Flag (lane 3), as indicated. Crude cell lysates were prepared and subjected to RNA-IP using anti-Flag antibody. RNA extracts prepared from precipitates and total cell lysates were subjected to Northern blots, as indicated. (E) Cultured S2 cells were transfected with empty vector (lane 1), the plasmid expressing EGFP-Flag (lane 2), Dcr-2–Flag (lane 3), or Dcr-2ΔPAZ-Flag (lane 4), as indicated. Total RNA extracts were prepared and subjected to Northern blots, as indicated. NB, Northern blots; IB, immunoblots or Western blots; GFP, green fluorescent protein.
Taken together, we conclude that Dcr-2 directly interacts with Toll 3′UTR via the PAZ domain and that this interaction plays a critical role in Toll signaling.
Dcr-2 is involved in Toll signaling in Drosophila S2 cells during viral infection
Previous studies reported that Toll immune signaling could be induced by, and/or could exert an antiviral effect on, RNA viruses such as Drosophila X virus, DCV, dengue virus, FHV, cricket paralysis virus, and Nora virus in insects (23, 25, 26, 31). Because we determined in this study that Dcr-2 is required for Toll immune signaling (Figs. 1 and 2), we sought to evaluate the effects of Dcr-2 in Toll signaling on certain viral infections. For this purpose, we first assessed the ability of a positive-stranded RNA virus, FHV, and of a negative-stranded RNA virus, VSV, to induce Toll signaling in cultured S2 cells. Our results showed that FHV dramatically induced Dros transcription 6 hours after infection (fig. S4A) whereas both FHV and VSV induced Dros transcription 48 hours after infection (fig. S4B), showing that Toll signaling can be activated by FHV and VSV in cultured S2 cells. Our additional experiments showed that Dcr-2 knockdown inhibited the Toll signaling induced by FHV (Fig. 8A, lane 7) or VSV (Fig. 8B, lane 7).
Fig. 8. Dcr-2 is involved in Toll signaling in S2 cells during FHV or VSV infection.
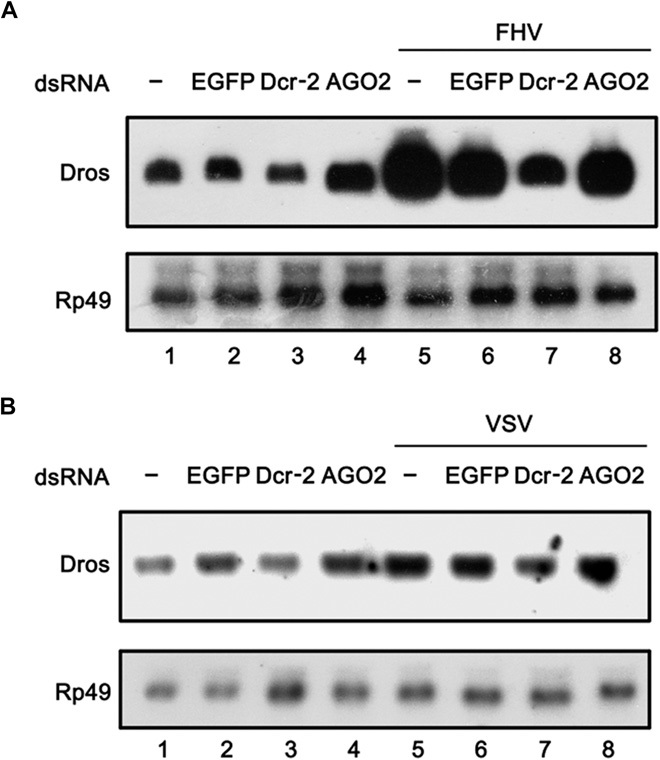
(A and B) Cultured S2 cells were transfected with dsRNAs against the indicated genes. The cells were mock-infected or infected with FHV (A) or VSV (B) for 6 hours. Total RNA extracts were prepared and subjected to Northern blots using the indicated probes.
DISCUSSION
In addition to its classic functions (RNA cleavage and gene silencing) in the RNAi pathway, Drosophila Dcr-2 has an RNAi-independent function that positively modulates the protein expression of Toll at the posttranscriptional level, consequently mediating Toll immune signaling. Our data further showed that Dcr-2 directly binds to the 3′UTR of Toll mRNA via its PAZ domain and is required for Toll 3′UTR–mediated protein expression, indicating that the direct interaction between Dcr-2 and Toll 3′UTR plays a critical role in the posttranscriptional modulation of Toll protein expression and Toll immune signaling.
In contrast to mammalian TLRs that function as PRRs to directly recognize pathogen-associated molecular patterns (PAMPs), Drosophila Toll is not a PRR and cannot be directly activated by bacteria or fungi. Gram-positive bacterial or fungal envelope PGNs are recognized as PAMPs by extracellular PGN recognition proteins such as PGRP-SA, PGRP-SD, GNBP-1, and GNBP-3, leading to cleavage of Spaetzle, which in turn activates Toll (27, 32–34). However, the mechanism by which viruses activate Toll immune signaling remains elusive. Dcr-2 is a cytoplasmic PRR that directly recognizes viral replicative intermediate dsRNAs in antiviral RNAi (14, 19). Given our current findings, this coincidence leads us to speculate that Dcr-2 acts as a viral sensor for Toll immune signaling and that the novel link between the RNAi pathway and the Toll pathway represents a plausible synergy of these two important immune pathways.
For Dicer proteins, the PAZ domain (also conserved in AGO proteins) binds to dsRNA or siRNA at 2-nucleotide overhangs on the 3′ end, which are generated by the Dicer-mediated cleavage of dsRNA (35, 36). Moreover, in vitro data of the Drosophila AGO PAZ domain revealed that the PAZ domain can interact with either single-stranded RNA or dsRNA (37). In this study, we uncovered that, in addition to its canonical dsRNA binding and cleavage activity, Drosophila Dcr-2 is also a novel 3′UTR binding protein that directly interacts with the 3′UTR of Toll mRNA. Indeed, although mRNA is single-stranded, most 3′UTR binding proteins bind to structured RNA elements that usually constitute short base pairings and single-stranded loops. Our prediction of Toll 3′UTR secondary structure, using two different algorithms of the RNAfold software, shows that Toll 3′UTR contains a number of structured RNA elements (fig. S5) that are not conventional substrates (that is, long dsRNA) of Dcr-2 in the RNAi pathway but probably interact with the PAZ domain of Dcr-2 that binds short single-stranded overhangs and RNA duplexes. This unconventional interaction between Dcr-2 and Toll 3′UTR should not support RNA cleavage by the RNase III activity of Dcr-2, consistent with our own observations.
The 3′UTRs of mRNAs contain a variety of posttranscriptional regulatory elements for protein expression. In eukaryotes, a number of eukaryotic initiation factors (eIFs) bind to the 5′UTR and interact with poly(A) binding protein, which binds to the poly(A) tail of the 3′UTR of mRNA. These protein-protein and protein-RNA interactions result in mRNA pseudocircularization, which is required for translation initiation. So far, most known 3′UTR binding proteins have been found to be repressive on protein expression via different strategies such as disrupting mRNA pseudocircularization, interfering with ribosome recruitment, shortening the poly(A) tail of mRNA, and promoting microRNA (miRNA)–mediated silencing (38). On the other hand, a few 3′UTR binding proteins positively modulate protein expression. For example, the mammalian eIF4E isoform 4EHP can activate translation under hypoxic conditions (39) even though the activating mechanism is unknown, and Dnd1 (dead end 1) can antagonize miRNA-mediated translational repression by competing with miRNA for 3′UTR binding (40). As a novel 3′UTR binding protein, Dcr-2 positively modulates Toll protein expression at the posttranscriptional level, which could be governed by diverse mechanisms such as antagonizing repressive 3′UTR binding proteins and/or miRNAs, enhancing ribosome recruitment, promoting the formation or stability of mRNA pseudocircularization, and stabilizing the poly(A) tail of mRNA. In addition, although the PAZ domain is responsible for Dcr-2 binding to Toll 3′UTR, other domains of Dcr-2 could also play a role in regulating Toll protein expression, probably by recruiting or repressing other cellular factors. In support of this idea, our data showed that the ectopic expression of a Flag-tagged PAZ domain repressed Toll 3′UTR–mediated protein expression and Toll signaling (Fig. 7, A and C), indicating that simply binding a PAZ domain to Toll 3′UTR is not enough to promote Toll signaling. It would be very interesting to elucidate in future studies the detailed mechanism by which Dcr-2 posttranscriptionally modulates protein expression.
Previous studies have provided several lines of evidence that the key RNase III–like enzymes of the RNAi machinery, Drosha and Dicer, have additional functions independent of their canonical roles (RNA cleavage and gene silencing) in RNAi. Drosha, the RNase III–like enzyme that processes primary miRNAs into pre-miRNAs, has been reported to bind promoter-proximal regions of many genes and to positively modulate transcription independently of RNAi (41). Moreover, Drosophila Dcr-2 and human Dicer have been associated with chromatin and have been reported to interact with the core transcription machinery, which may regulate transcription by affecting RNA polymerase II processivity (42, 43). In addition, Dcr-2 has been reported to mediate the virus-induced induction of Vago (20). However, the mechanism by which Dcr-2 affects Vago induction seems be different from the mechanism by which Dcr-2 affects Toll signaling because Dcr-2–mediated Vago induction occurs independently of the Toll pathway. Besides, Vago induction also occurs independently of the IMD pathway (20).
To our knowledge, this study was the first to provide evidence that a Dicer protein positively modulates protein expression at the posttranscriptional level instead of mediating posttranscriptional gene silencing. Moreover, our findings also revealed an unexpected link between the RNAi pathway and the Toll pathway. Given the high degree of homology among Dicer and Dicer-related proteins in different organisms, including plants, invertebrates, and mammals, this study opens up the possibility that this protein group functions as a novel class of posttranscriptional regulators of gene expression in diverse pathways, such as innate immunity and various other processes, and represents an exciting direction toward future research.
MATERIALS AND METHODS
Fly stocks and microbial challenge
Three- to 5-day-old adult flies were reared at 25°C and fed a standard cornmeal/yeast diet. Adult flies were randomly allocated, and the sample size was chosen based on a previous study, Galiana-Arnoux et al. (19). The Actin-GAL4/CyO-PscGFP driver line was obtained from N. Dostatni (Laboratory of Nuclear Dynamics and Genome Plasticity, Paris, France). UAS-dsRNA (Dcr-2), Dcr-2L811fsX, and r2d21 fly lines were provided by Q.Wu (University of Science and Technology of China, Hefei, China). The Ago2414 fly line was obtained from the Bloomington Stock Center. The yw fly line used for dsRNA injection and in controls was obtained from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences (Beijing, China).
M. luteus was provided by F. Peng (China Center for Type Culture Collection, Wuhan, China). For the M. luteus challenge, adult flies (3 to 5 days old) were inoculated by a needle previously dipped into a concentrated culture of M. luteus. A. fumigatus was provided by Z. Liu (Huazhong Agricultural University, Wuhan, China). For A. fumigatus infection, adult flies (3 to 5 days old) were inoculated by a needle previously dipped into a concentrated solution of A. fumigatus spores.
Plasmid and in vitro transcription of RNA or dsRNA
The Drosophila-inducible expression system vector pMT/V5-His (Invitrogen) was used to construct plasmids that express proteins in Drosophila S2 cells. dcr-2 or Toll ORF was amplified from fly cDNAs (provided by J. Ni, Tsinghua University, Beijing, China) via PCR. The dcr-2 ORF or its mutant carrying a Flag tag at its 3′ end was cloned into the Kpn І–Not І site of the pMT/V5-His vector downstream of the copper sulfate–inducible metallothionein promoter. The Toll ORF carrying a Flag tag at its 5′ end was cloned into the Kpn І–Xba І site of the pMT/V5-His vector downstream of the copper sulfate–inducible metallothionein promoter. During transfection, 500 μM copper sulfate was added into the standard S2 medium. The PAZ domain and the dsRBD carrying a Flag tag at its 3′ end were cloned into the Eco RІ–Xba І site of the pAc5.1/V5-His B vector (Invitrogen) downstream of the Drosophila Actin 5C promoter.
The 8×His-tagged Dcr-2 recombinant protein was expressed in insect cells using the Bac-to-Bac Baculovirus Expression System. The dcr-2 ORF carrying an N-terminal 8×His tag was cloned into the Nco І–Not І site of the pFastBac1 vector (Invitrogen). The expression vector pMAL-c2x (New England Biolaboratories) was used to construct plasmids that express the PAZ domain, dsRBD, and RNase III domain of Dcr-2 in Escherichia coli. The corresponding coding sequences were amplified from fly cDNAs by PCR and cloned into the Eco RІ–Sal І (PAZ domain and dsRBD) or Eco RІ–Hind III (RNase III domain) site of pMAL-c2x. The vector pGEX-6P-1 (GE Healthcare) was used to construct plasmids that express the DExD/H-box helicase domain of Dcr-2 in E. coli. The corresponding coding sequence was amplified from fly cDNAs by PCR and cloned into the Eco RІ–Xho I site of pGEX-6P-1.
dsRNAs used for RNAi and exogenous Toll 3′UTR RNA were transcribed in vitro from PCR products for 4 hours using T7 RNA polymerase (Promega). The primers used for PCR amplification are listed in table S1.
Expression and purification of recombinant proteins
The recombinant 8×His-tagged Dcr-2 was expressed and purified using the Bac-to-Bac Baculovirus Expression System according to our standard protocol (44). In brief, recombinant baculovirus-infected Sf9 cells were collected and lysed using glass beads in a Retsch MM400 machine (30 Hz/30 s). The cell lysates were loaded into a 2-ml nickel column in phosphate-buffered saline (PBS; 300 mM NaCl) with 1× Protease Inhibitor Cocktail (Roche). After a wash with PBS and PBS + 10 mM imidazole, we eluted the column with 10 ml of PBS + 300 mM imidazole. Nickel elution was concentrated to 1 ml and diluted to 10 ml with NaCl-free buffer A [10 mM Hepes (pH 7.4), 10 mM KOAc, 2 mM Mg(OAc)2, and 5 mM β-mercaptoethanol]. The samples were centrifuged at 15,000 rpm for 5 min and loaded into a MonoQ column. We performed gradient elution using 0 to 60% buffer B [1 M NaCl, 10 mM Hepes (pH 7.4), 10 mM KOAc, 2 mM Mg(OAc)2, and 5 mM β-mercaptoethanol] and wash using 100% buffer B. His8–Dcr-2 elution peaked with ~30% buffer B.
MBP-tagged recombinant proteins were expressed and purified according to our standard protocol (45). Protein expression was induced by 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested and resuspended using MBP lysis buffer [20 mM tris-HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, 1 mM sodium azide, 10 mM β-mercaptoethanol, and 1 mM dithiothreitol (DTT)] after incubation at 30°C for 6 hours. The soluble recombinant proteins were purified using amylose resin (New England Biolaboratories) according to the manufacturer’s protocol. Glutathione S-transferase (GST)–tagged recombinant protein was expressed and purified according to our standard protocol, Qi et al. (46). Protein expression was induced by 0.8 mM IPTG. Cells were harvested and resuspended using 1× PBS [1.4 M NaCl, 27 mM KCl, 100 mM Na2HPO4, and 18 mM KH2PO4 (pH 7.5)] after incubation at 25°C for 6 hours. The soluble recombinant proteins were purified using Glutathione Sepharose 4B (GE Healthcare) according to the manufacturer’s protocol. The purified proteins were concentrated using Amicon Ultra-15 filters (Millipore) and stored in 25 mM Hepes (pH 7.5) at −20°C.
Cell culture and transfection
The cell lines used in this study were obtained from the China Center for Type Culture Collection. Drosophila S2 cells were grown and maintained in Schneider’s Insect Medium (Sigma)/10% fetal bovine serum (FBS) and penicillin/streptomycin, as previously described, Qi et al. (47). Sf9 cells were grown and maintained in Grace’s Insect Medium (Invitrogen)/10% FBS and penicillin/streptomycin. Baby hamster kidney 21 (BHK21) cells were maintained in Dulbecco’s modified Eagle’s medium/10% FBS and penicillin/streptomycin.
DNA or RNA transfection was performed as previously described, Qui et al. (48). In brief, Drosophila S2 cells were plated in six-well plates and grown overnight to reach 80% confluence (about 106 cells per well). Two micrograms of DNA plasmid or 1 μg of RNA was transfected into the cells using FuGENE HD Transfection Reagent (Roche), according to the manufacturer’s protocol. For PGN or virus challenge, the cells were challenged by Lys-PGN [extract from S. aureus (Sigma)] or infected with the indicated viruses 72 hours after the transfection of dsRNA.
Western blots and antibodies
Cultured Drosophila S2 cells were harvested and lysed in cell lysis buffer. The cell lysates were subjected to 10% SDS–polyacrylamide gel electrophoresis (PAGE) and Western blots according to our standard procedures (45). Anti-Flag M2 monoclonal antibody (F1804; Sigma) was used at a dilution of 1:2000. Anti–α-tubulin monoclonal antibody (66031-1-Ig; ProteinTech) was used at a dilution of 1:3000. Anti-Toll polyclonal antibody (sc-15693; Santa Cruz Biotechnology) was used at a dilution of 1:250. Anti–green fluorescent protein polyclonal antibody (cw0087; CWbio) was used at a dilution of 1:3000.
Northern blots, RT-PCR, and qRT-PCR
Total RNA was extracted from cells using TRIzol reagent (TaKaRa Bio) and treated with RQ1 RNase-free DNase I (Promega) to remove DNAs, as previously described, Qi et al. (46). Northern blots were performed according to our standard procedures (48).
For RT-PCR, RNAs extracted from cells were subjected to reverse transcription with M-MLV Reverse Transcriptase (Promega) using random primers, according to our standard procedures (45). qRT-PCR was performed using a SuperReal PreMix Plus kit (Tiangen), according to the manufacturer’s protocol. Gene-specific primers used for PCR amplification or qRT-PCR are listed in table S1.
IP and RNA-IP assays
IP assays were conducted according to our standard protocol (46). Proteins were extracted from precipitates and subjected to 10% SDS-PAGE and Western blots.
For RNA-IP assays, transfected S2 cells were lysed in RNA-IP lysis buffer [150 mM KCl, 25 mM tris (pH 7.4), 5 mM EDTA, 0.5 mM DTT, 0.5% NP-40, 100 U/ml RNase inhibitor, and 1× Protease Inhibitor Cocktail (Roche)]. The cell lysates were incubated with anti-Flag M2 monoclonal antibody (F1804; Sigma) at 4°C for 6 hours. Twenty microliters of Protein A/G Agarose (Santa Cruz Biotechnology) were supplemented and incubated at 4°C for 1 hour. After three washes, RNAs were extracted from the precipitates using TRIzol reagent (TaKaRa Bio). The extracted RNAs were detected by Northern blots or RT-PCR, as previously described.
Gel shift assay
Gel shift assays were performed according to our standard protocol (46). Purified 8×His-tagged full-length Dcr-2, MBP, MBP-tagged PAZ domain, MBP-tagged dsRBD, MBP-tagged RNase III domain, GST, or GST-tagged DExD/H-box helicase domain at a concentration of 30 μM was incubated with 0.1 μM in vitro–transcribed DIG-labeled Toll 3′UTR at 25°C for 30 min. After incubation, the reaction mixture was mixed with 6× RNA loading buffer and subjected to electrophoresis with 1.5% nondenaturing agarose gel. The reaction products were transferred onto a N+ nylon membrane (Millipore) and fixed. The nylon membrane was treated with anti–DIG-AP antibody (Roche) and incubated with CDP-Star (Roche) at 37°C for 10 to 15 min. The signal was visualized by radiography on an x-ray film according to our standard protocol (45).
Viruses
Full-length infectious cDNA clones for FHV RNAs 1 and 2 were obtained from S.-W. Ding (University of California, Riverside, CA). FHV was extracted from S2 cells transfected with both of the FHV cDNA clones, as previously described, Wang et al. (14). VSV was provided by H.-B. Shu (Wuhan University, Wuhan, China) and grown in BHK21 cells, as previously described, Zhong et al. (49).
Acknowledgments
We thank J. Ni (Beijing, China) for fly cDNAs; S.-W. Ding (Riverside, CA) for FHV clones; H.-B. Shu (Wuhan, China) for VSV; Q. Wu (Hefei, China) for UAS-dsRNA (Dcr-2), Dcr-2L811fsX, and r2d21 fly lines; F. Peng (Wuhan, China) for M. luteus; and Z. Liu (Wuhan, China) for A. fumigatus. We also thank S.-W. Ding (Riverside, CA), X. Chen (Riverside, CA), and Y. Yu (Cold Spring Harbor, NY) for helpful discussions, and M. Wade (Houston, TX) for professionally editing the manuscript. Funding: This work was supported by the National High-Tech R&D Program of China (863 Program; grant 2015AA020939 to X.Z.), the National Basic Research Program of China (973 Program; grant 2014CB542603), the National Natural Science Foundation Excellent Young Scientists Fund (grant 31522004 to X.Z.), the National Natural Science Foundation of China (grants 31270190 and 81201292 to X.Z.), the Fundamental Research Funds for the Central Universities (grant 204274386 to Z.W.), and the Chinese 111 Project (grant B06018). Author contributions: Z.W. and X.Z. conceived and designed the experiments; Z.W., D.W., Y.L., X.X., W.G., Y.Q., J.Y., Y.Z., J.L., Y.F.-W., and Y.X. performed the experiments; Z.W., Y.H., and X.Z. analyzed the data; and Z.W. and X.Z. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the findings in the paper are present in the paper and Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500228/DC1
Fig. S1. Overexpression of Dcr-2 enhances Toll signaling activation in Drosophila S2 cells.
Fig. S2. Toll 3′UTR is important for Toll signaling.
Fig. S3. The PAZ domain of Dcr-2 has no interaction with the 3′UTR of DIF or Dorsal in vitro.
Fig. S4. Toll signaling can be induced by FHV or VSV.
Fig. S5. The prediction of the secondary structure of Toll 3′UTR.
Table S1. Primers used in this work.
REFERENCES AND NOTES
- 1.Hoffmann J. A., The immune response of Drosophila. Nature 426, 33–38 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Xu J., Cherry S., Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 42, 67–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kemp C., Imler J.-L., Antiviral immunity in Drosophila. Curr. Opin. Immunol. 21, 3–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govind S., Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 15, 29–43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng X., Khanuja B. S., Ip Y. T., Toll receptor–mediated Drosophila immune response requires Dif, an NF-κB factor. Genes Dev. 13, 792–797 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hetru C., Hoffmann J. A., NF-κB in the immune response of Drosophila. Cold Spring Harb. Perspect. Biol. 1, a000232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khush R. S., Leulier F., Lemaitre B., Drosophila immunity: Two paths to NF-κB. Trends Immunol. 22, 260–264 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Bulet P., Hetru C., Dimarcq J.-L., Hoffmann D., Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 23, 329–344 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Ferrandon D., Jung A. C., Criqui M.-C., Lemaitre B., Uttenweiler-Joseph S., Michaut L., Reichhart J.-M., Hoffmann J. A., A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17, 1217–1227 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samakovlis C., Kimbrell D. A., Kylsten P., Engström A., Hultmark D., The immune response in Drosophila: Pattern of cecropin expression and biological activity. EMBO J. 9, 2969–2976 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz D. S., Hutvágner G., Du T., Xu Z., Aronin N., Zamore P. D., Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Hutvagner G., Simard M. J., Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Tomari Y., Zamore P. D., Perspective: Machines for RNAi. Genes Dev. 19, 517–529 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Wang X. H., Aliyari R., Li W. X., Li H. W., Kim K., Carthew R., Atkinson P., Ding S. W., RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q., Rand T. A., Kalidas S., Du F., Kim H. E., Smith D. P., Wang X., R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301, 1921–1925 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Marques J. T., Kim K., Wu P.-H., Alleyne T. M., Jafari N., Carthew R. W., Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 17, 24–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Lu J., Han Y., Fan X., Ding S. W., RNA interference functions as an antiviral immunity mechanism in mammals. Science 342, 231–234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maillard P. V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S. W., Voinnet O., Antiviral RNA interference in mammalian cells. Science 342, 235–238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galiana-Arnoux D., Dostert C., Schneemann A., Hoffmann J. A., Imler J.-L., Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nat. Immunol. 7, 590–597 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Deddouche S., Matt N., Budd A., Mueller S., Kemp C., Galiana-Arnoux D., Dostert C., Antoniewski C., Hoffmann J. A., Imler J.-L., The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in Drosophila. Nat. Immunol. 9, 1425–1432 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T., The RNA helicase RIG-I has an essential function in double-stranded RNA–induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Zhang H. X., Liu Z.-X., Sun Y.-P., Zhu J., Lu S.-Y., Liu X.-S., Huang Q.-H., Xie Y. Y., Zhu H.-B., Dang S.-Y., Chen H.-F., Zheng G.-Y., Li Y.-X., Kuang Y., Fei J., Chen S.-J., Chen Z., Wang Z.-G., Rig-I regulates NF-κB activity through binding to Nf-κb1 3′-UTR mRNA. Proc. Natl. Acad. Sci. U.S.A. 110, 6459–6464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zambon R. A., Nandakumar M., Vakharia V. N., Wu L. P., The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 102, 7257–7262 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaitre B., Reichhart J.-M., Hoffmann J. A., Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. U.S.A. 94, 14614–14619 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez J. L., Dimopoulos G., The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev. Comp. Immunol. 34, 625–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xi Z., Ramirez J. L., Dimopoulos G., The Aedes aegypti Toll pathway controls dengue virus infection. PLOS Pathog. 4, e1000098 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leulier F., Parquet C., Pili-Floury S., Ryu J.-H., Caroff M., Lee W.-J., Mengin-Lecreulx D., Lemaitre B., The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4, 478–484 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Lemaitre B., Nicolas E., Michaut L., Reichhart J.-M., Hoffmann J. A., The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Huang H.-R., Chen Z. J., Kunes S., Chang G.-D., Maniatis T., Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 8322–8327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor D. W., Ma E., Shigematsu H., Cianfrocco M. A., Noland C. L., Nagayama K., Nogales E., Doudna J. A., Wang H.-W., Substrate-specific structural rearrangements of human Dicer. Nat. Struct. Mol. Biol. 20, 662–670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira Á. G., Naylor H., Esteves S. S., Pais I. S., Martins N. E., Teixeira L., The Toll-dorsal pathway is required for resistance to viral oral infection in Drosophila. PLOS Pathog. 10, e1004507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gobert V., Gottar M., Matskevich A. A., Rutschmann S., Royet J., Belvin M., Hoffmann J. A., Ferrandon D., Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science 302, 2126–2130 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Bischoff V., Vignal C., Boneca I. G., Michel T., Hoffmann J. A., Royet J., Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 5, 1175–1180 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Ligoxygakis P., Pelte N., Hoffmann J. A., Reichhart J. M., Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297, 114–116 (2002). [DOI] [PubMed] [Google Scholar]
- 35.MacRae I. J., Zhou K., Doudna J. A., Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 14, 934–940 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Lingel A., Simon B., Izaurralde E., Sattler M., Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 11, 576–577 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Yan K. S., Yan S., Farooq A., Han A., Zeng L., Zhou M.-M., Structure and conserved RNA binding of the PAZ domain. Nature 426, 468–474 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Szostak E., Gebauer F., Translational control by 3′-UTR-binding proteins. Brief. Funct. Genomics 12, 58–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho P. F., Poulin F., Cho-Park Y. A., Cho-Park I. B., Chicoine J. D., Lasko P., Sonenberg N., A new paradigm for translational control: Inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121, 411–423 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A. F., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., van Duijse J., Ørom U. A., Lund A. H., Perrakis A., Raz E., Agami R., RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273–1286 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Gromak N., Dienstbier M., Macias S., Plass M., Eyras E., Cáceres J. F., Proudfoot N. J., Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 5, 1499–1510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White E., Schlackow M., Kamieniarz-Gdula K., Proudfoot N. J., Gullerova M., Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat. Struct. Mol. Biol. 21, 552–559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cernilogar F. M., Onorati M. C., Kothe G. O., Burroughs A. M., Parsi K. M., Breiling A., Sardo F. L., Saxena A., Miyoshi K., Siomi H., Siomi M. C., Carninci P., Gilmour D. S., Corona D. F. V., Orlando V., Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480, 391–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., Han Y., Qiu Y., Zhang S., Tang F., Wang Y., Zhang J., Hu Y., Zhou X., Identification and characterization of RNA duplex unwinding and ATPase activities of an alphatetravirus superfamily 1 helicase. Virology 433, 440–448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Qiu Y., Liu Y., Qi N., Si J., Xia X., Wu D., Hu Y., Zhou X., Characterization of a nodavirus replicase revealed a de novo initiation mechanism of RNA synthesis and terminal nucleotidyltransferase activity. J. Biol. Chem. 288, 30785–30801 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi N., Cai D., Qiu Y., Xie J., Wang Z., Si J., Zhang J., Zhou X., Hu Y., RNA binding by a novel helical fold of B2 protein from wuhan nodavirus mediates the suppression of RNA interference and promotes B2 dimerization. J. Virol. 85, 9543–9554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi N., Zhang L., Qiu Y., Wang Z., Si J., Liu Y., Xiang X., Xie J., Qin C.-F., Zhou X., Hu Y., Targeting of dicer-2 and RNA by a viral RNA silencing suppressor in Drosophila cells. J. Virol. 86, 5763–5773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu Y., Cai D., Qi N., Wang Z., Zhou X., Zhang J., Hu Y., Internal initiation is responsible for synthesis of Wuhan nodavirus subgenomic RNA. J. Virol. 85, 4440–4451 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong B., Yang Y., Li S., Wang Y.-Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H.-B., The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/9/e1500228/DC1
Fig. S1. Overexpression of Dcr-2 enhances Toll signaling activation in Drosophila S2 cells.
Fig. S2. Toll 3′UTR is important for Toll signaling.
Fig. S3. The PAZ domain of Dcr-2 has no interaction with the 3′UTR of DIF or Dorsal in vitro.
Fig. S4. Toll signaling can be induced by FHV or VSV.
Fig. S5. The prediction of the secondary structure of Toll 3′UTR.
Table S1. Primers used in this work.


