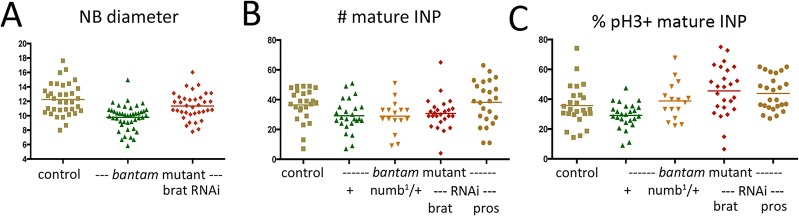
Fig. 5.
Genetic evidence that bantam acts via regulation of brat and prospero. (A-C) MARCM clonal analysis showing bantamΔ1/Δ1 mutant clones expressing the indicated UAS-RNAi transgenes. (A) Type II NB diameter (µm) in control FRT2A clones, bantamΔ1/Δ1 mutant clones and bantamΔ1/Δ1 mutant clones expressing UAS-brat RNAi. Control: n=35; bantam mutant n=42; with brat RNAi n=36 clones. ANOVA: P<0.0001 comparing control and bantam mutant; P=0.0008 comparing bantam mutant with and without brat RNAi. The control and bantam mutant samples are the same as those shown in Fig. 3E. NB diameter was not measured in the brat RNAi control clones in an otherwise wild-type background. (B) Total number of mature INPs (Dpn+Ase+) in type II NB clones of the indicated genotypes. n=25 clones for the control, bantamΔ1/Δ1 mutant and bantam mutant with brat RNAi. n=34 clones for the bantam mutant with prospero RNAi and n=16 with numb1. ANOVA: P=0.029 comparing bantam mutant with and without prospero RNAi. The other comparisons were not significant. The control and bantam mutant samples are the same as those shown in Fig. 3G. Each clone contained a single large Dpn+Ase− NB, so the change in clone size and INP number/clone cannot be attributed to an increase in NB number, as occurs in brat or prospero mutant brains. (C) Percentage of mature INPs labeled with anti-pH3 in type II NB clones of the indicated genotypes. n=25 clones for the control, bantamΔ1/Δ1 mutant and bantam mutant with brat or prospero RNAi and n=16 with numb1. ANOVA: P<0.0001 comparing bantam mutant with and without brat RNAi and P=0.0005 comparing bantam mutant with/without prospero RNAi. The effect of removing one copy of numb was not statistically significant by ANOVA (P=0.07, comparing all samples in the experiment), but was significant in a pairwise comparison of the bantam mutant with and without numb RNAi using an unpaired t-test (P=0.013 assuming unequal variance). The control and bantam mutant samples are the same as those shown in Fig. 3J.