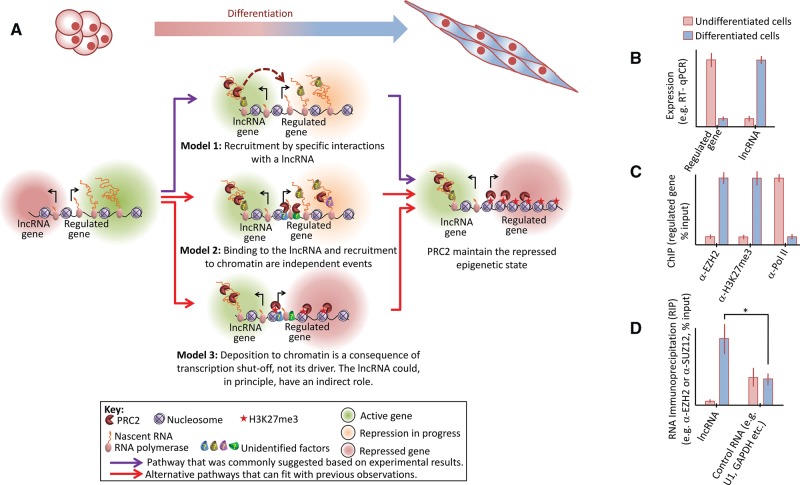
FIGURE 3.
Alternative explanations for observations that were previously provided to support the recruitment of PRC2 for epigenetic repression through specific interactions with lncRNAs. (A) In undifferentiated cells the differentially regulated gene is active and the lncRNA is repressed or lowly expressed. The cascade of events that could take place through differentiation is illustrated by three models, all of which are compatible with current literature. Model 1 (top, purple arrows): A given lncRNA is differentially expressed and binds PRC2 by specific protein–RNA interactions, either directly or through a bridging factor. This leads to the recruitment of PRC2 to the regulated gene (dashed arrow). Next, PRC2 introduces the H3K27me3 mark. Model 2 (middle, red arrows): The lncRNA is differentially expressed and also binds PRC2, possibly through promiscuous protein–RNA interactions. Yet, the association of PRC2 with the lncRNA does not directly cause its deposition to chromatin. Instead, recruitment of PRC2 to the regulated gene takes place independently by other factors (for reviews, see Ringrose and Paro 2007; Margueron and Reinberg 2011; Di Croce and Helin 2013; Simon and Kingston 2013; Comet and Helin 2014). Model 3 (bottom, red arrows): The differentially regulated gene is repressed in a PRC2-independent manner. The lncRNA could function in this process, but it is not the driving force for the recruitment of PRC2, which only interacts with the RNA through promiscuous interactions. Next, transcription shutoff leads to the recruitment of PRC2 to chromatin (Riising et al. 2014), where it maintains the repressed epigenetic state. In all three models, the end point is identical: In differentiated cells the lncRNA is expressed while the differentially regulated gene is epigenetically repressed, with PRC2 maintaining the repressed epigenetic state. (B–D) Hypothetical in vivo data commonly provided to support the recruitment of PRC2 by specific interactions with lncRNAs (see text for details). (B) Expression analysis supports all three models, but cannot exclude any of them. (C) Chromatin immunoprecipitation (ChIP) for the differentially expressed gene identifies the recruitment of PRC2 to chromatin during differentiation (α-EZH2), deposition of its repressive epigenetic mark (α-H3K27me3), and reduction in RNA polymerase occupancy over the regulated gene (α-Pol II), yet these observations agree with all three models and are therefore not definitive. (D) RNA immunoprecipitation confirms that PRC2 is associated with the lncRNA to a greater extent, compared with a negative control RNA, to a statistically significant degree. Although this observation is consistent with Model 1, it is not definitive and can also be explained by an undetected bridging factor or simply by a higher degree of promiscuous interactions, as previously observed between PRC2 and various nonrelevant RNAs in vivo (Kaneko et al. 2013) and suggested to be the result of competition between PRC2 and other RNA-binding proteins (Herzog et al. 2014).