FIGURE 2.
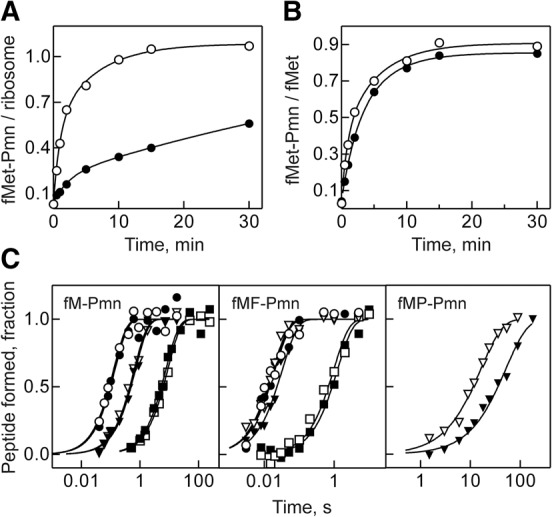
Peptide-bond formation with Pmn as A-site substrate. Unpurified (A) or sucrose gradient-purified (B) ICs prepared with wt (open circles) or ΔL27 (closed circles) ribosomes (0.25 µM) were mixed with Pmn (1 µM), and the extent of dipeptide reaction was monitored over time. Solid lines represent the results of exponential fitting (see Materials and Methods). (C) Time courses of fMet-Pmn (left panel), fMetPhe-Pmn (middle panel), and fMetPro-Pmn (right panel) formation upon mixing of wt (open symbols) and ΔL27 (closed symbols) IC (50 nM) with high concentrations of Pmn (2.5–10 mM) at pH 6.5 (squares), 7.5 (triangles), and 8.5 (circles). Time courses were normalized for the extent of the reaction to facilitate visual inspection. Solid lines represent the results of exponential fitting of the time points. Rates of the fMet-Pmn reaction for wt and ΔL27 ribosomes, respectively, were 0.11 ± 0.1 and 0.13 ± 0.1 sec−1 at pH 6.5; 1.6 ± 0.1 and 1.4 ± 0.1 sec−1 at pH 7.5; and 7.8 ± 1.7 and 7.1 ± 1.9 sec−1 at pH 8.5. Rates of the fMetPhe-Pmn for wt and ΔL27 ribosomes, respectively, were 1.0 ± 0.2 and 0.8 ± 0.1 sec−1 at pH 6.5; 55 ± 6 and 30 ± 3 sec−1 at pH 7.5; and 55 ± 10 and 49 ± 10 sec−1 at pH 8.5. Rates of the fMetPro-Pmn reaction at pH 7.5 were 0.06 ± 0.01 and 0.02 ± 0.01 sec−1 for wt and ΔL27 ribosome, respectively.
