FIGURE 4.
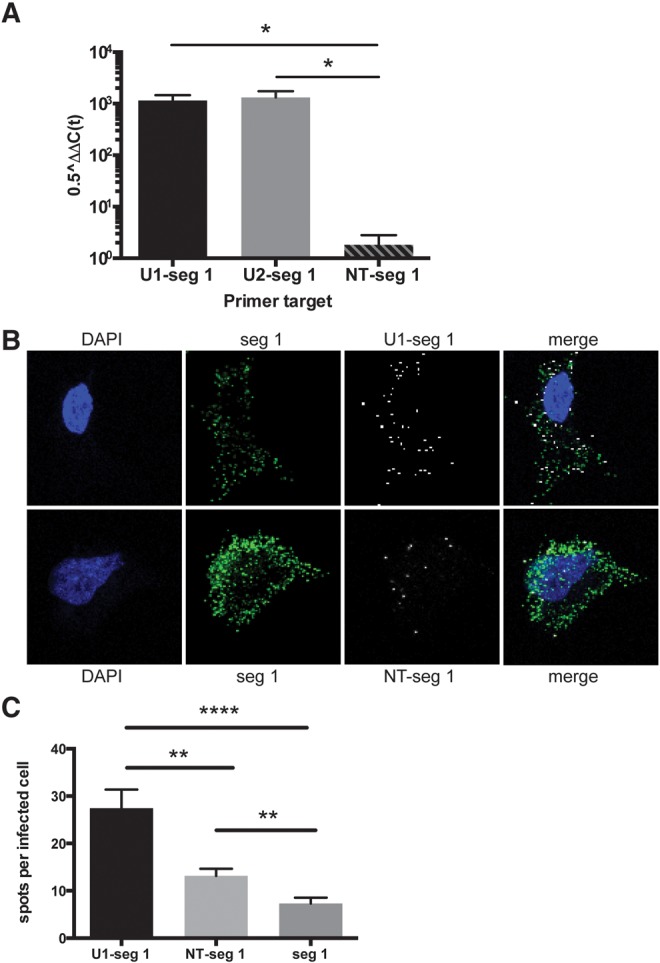
Verification of commonly snatched human RNA sequences by qRT-PCR and RNA fluorescence in situ hybridization (FISH). (A) qRT-PCR analysis was performed on samples from influenza virus–infected cells at 24 h. Host–virus hybrid products are shown as 0.5ΔΔCt normalized to host HPRT. Hybrids of U1-influenza segment 1 and U2-segment 1 are abundant compared with a nontargeting (NT) sequence hybrid, NT-segment 1. Error bars represent the SEM for three independent infection experiments. (*) P < 0.05; two-tailed t-test. (B) FISH was performed on virus-infected cells at 24 h. Blue is DAPI, green is a probe exclusively against segment 1, and white is a probe against the U1-segment 1 hybrid (top row) or an NT-segment 1 hybrid probe (bottom row). (C) Quantification of the number of hybrid probe spots per infected cell as identified by the segment 1 probe staining (n = 47, U1-seg 1; n = 55, NT–seg 1; n = 54, seg 1). Error bars represent the SEM. (**) P = 0.001; (****) P < 0.0001; two-tailed t-test.
