Fig. 2.
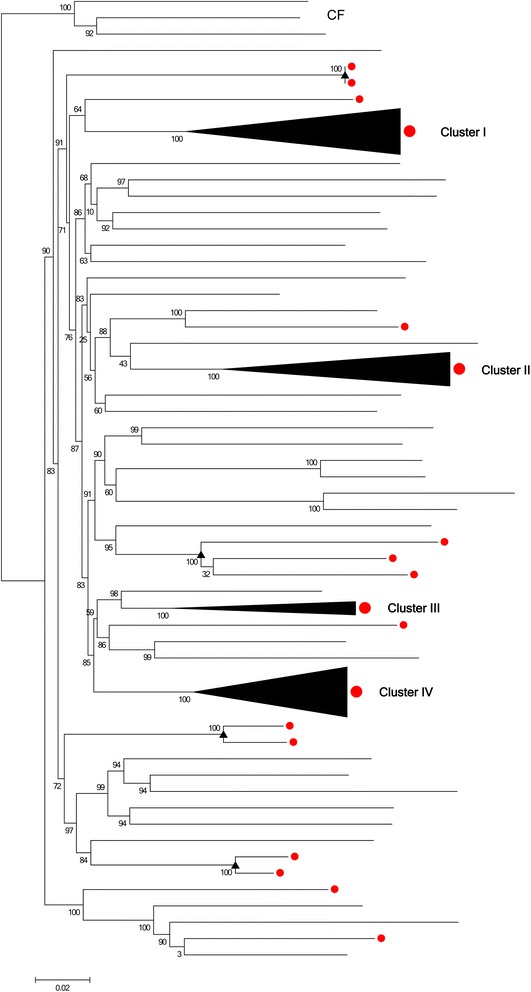
Recombinants located in the phylogenetic tree of NFLG HIV-1 CRF01_AE strains from China. Near-full-length genome (NFLG) sequences from China (n = 153) were analyzed in a maximum likelihood phylogenic tree. The tree was constructed using PhyML. SH values of all relevant nodes are indicated. SH support values of ≥90 % were here considered significant. A total of 84 CRF01_AE strains of 153 were found to be intrasubtype recombinants. All 84 sequences showing significant evidence (recombinants) are here labeled with red dots. Three large (≥10 sequences), well-supported (SH values were 100 %) and distinct clusters of CRF01_AE strains being almost entirely composed of putative recombinants were indicated from clusters I–III. Much smaller recombinant clusters including only 2 or 3 sequences are indicated by triangles. Cluster IV (SH value was 100 %) represents a nonrecombinant cluster. The outgroup is indicated with CF
